Installation
First, download and install R and RStudio:
Then, open RStudio and install the devtools package
install.packages("devtools")
Finally, install the MotrpacBicQC package
library(devtools) devtools::install_github("MoTrPAC/MotrpacBicQC", build_vignettes = TRUE)
Usage
Load the library
library(MotrpacBicQC)
And run any of the following tests to check that the package is correctly installed it works. For example:
# Just copy and paste in the RStudio terminal check_metadata_metabolites(df = metadata_metabolites_named, name_id = "named") check_metadata_samples(df = metadata_sample_named, cas = "umichigan") check_results(r_m = results_named, m_s = metadata_sample_named, m_m = metadata_metabolites_named)
which should generate the following output:
check_metadata_metabolites(df = metadata_metabolites_named, name_id = "named") #> + (+) All required columns present #> + (+) {metabolite_name} OK #> + (+) {refmet_name} unique values: OK #> + (+) {refmet_name} ids found in refmet: OK #> + (+) {rt} all numeric: OK #> + (+) {mz} all numeric: OK #> + (+) {neutral_mass} all numeric values OK #> + (+) {formula} available: OK check_metadata_samples(df = metadata_sample_named, cas = "umichigan") #> + (+) {sample_id} seems OK #> + (+) {sample_type} seems OK #> + (+) {sample_order} is numeric #> + (+) {sample_order} unique values OK #> + (+) {raw_file} unique values OK check_results(r_m = results_named, m_s = metadata_sample_named, m_m = metadata_metabolites_named) #> + (+) All samples from [results_metabolite] are available in [metadata_sample] #> + (+) {metabolite_name} is identical in both [results] and [metadata_metabolites] files: OK #> + (+) {sample_id} columns are numeric: OK
How to test your datasets
Two approaches available:
Check full PROCESSED_YYYYMMDD folder (recommended)
Run test on the full submission. For that, run the following command:
validate_metabolomics(input_results_folder = "/full/path/to/PROCESSED_YYYYMMDD", cas = "your_site_code")
cas is one of the followings:
- “broad_met” = Broad Metabolomics
- “emory” = Emory
- “mayo” = Mayo Clinic
- “umichigan” = Umichigan
- “gtech” = Georgia Tech
- “duke” = Duke
Check individual files
- Check metadata metabolites:
# Open the metadata_metabolites file(s) metadata_metabolites_named <- read.delim(file = "/path/to/your/file", stringsAsFactors = FALSE) metadata_metabolites_unnamed <- read.delim(file = "/path/to/your/file", stringsAsFactors = FALSE) check_metadata_metabolites(df = metadata_metabolites_named, name_id = "named") check_metadata_metabolites(df = metadata_metabolites_unnamed, name_id = "unnamed")
- Check metadata samples:
# Open your files metadata_sample_named <- read.delim(file = "/path/to/your/file", stringsAsFactors = FALSE) metadata_sample_unnamed <- read.delim(file = "/path/to/your/file", stringsAsFactors = FALSE) check_metadata_samples(df = metadata_sample_named, cas = "your_side_id") check_metadata_samples(df = metadata_sample_unnamed, cas = "your_side_id")
- Check results, which needs both both metadata metabolites and samples
# Open your files metadata_metabolites_named <- read.delim(file = "/path/to/your/file", stringsAsFactors = FALSE) metadata_sample_named <- read.delim(file = "/path/to/your/file", stringsAsFactors = FALSE) results_named <- read.delim(file = "/path/to/your/file", stringsAsFactors = FALSE) check_results(r_m = results_named, m_s = metadata_sample_named, m_m = metadata_metabolites_named)
Merge metabolomics data (only PASS1A supported)
The following functions enable merging all results and metadata files into a single data frame.
The folder/file structure of a required untargeted metabolomics submission is as follows:
PASS1A-06/
T55/
HILICPOS/
BATCH1_20190725/
RAW/
Manifest.txt
file1.raw
file2.raw
etc
PROCESSED_20190725/
metadata_failedsamples_[cas_specific_labeling]. txt
NAMED/
results_metabolites_named_[cas_specific_labeling].txt
metadata_metabolites_named_[cas_specific_labeling].txt
metadata_sample_named_[cas_specific_labeling].txt
metadata_experimentalDetails_named_[cas_specific_labeling].txt
UNNAMED/
results_metabolites_unnamed_[cas_specific_labeling].txt
metadata_metabolites_unnamed_[cas_specific_labeling].txt
metadata_sample_unnamed_[cas_specific_labeling].txt
metadata_experimentalDetails_unnamed_[cas_specific_labeling].txtWith the following file relations…
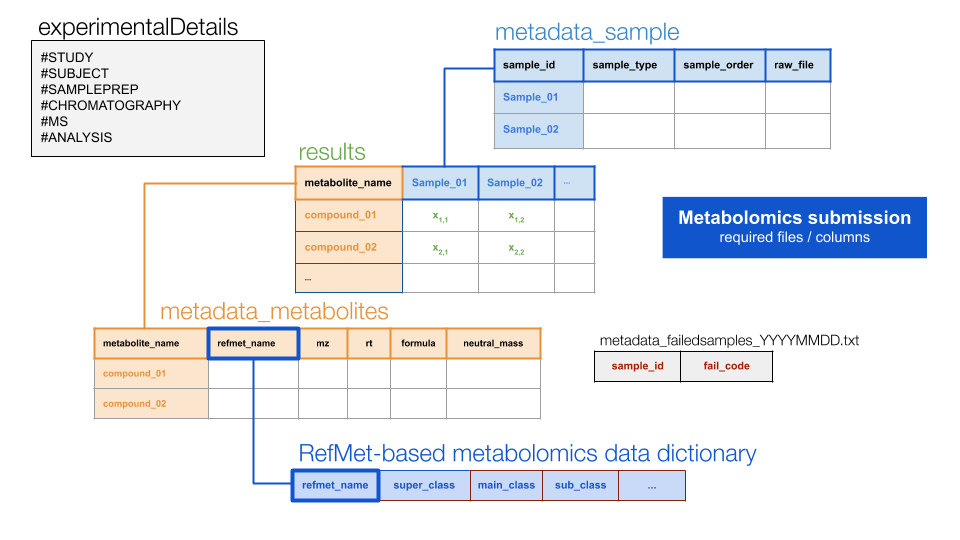
To merge all data available in a PROCESSED_YYYYMMDD folder, run the following command:
t31_ionpneg <- combine_metabolomics_batch(input_results_folder = "/full/path/to/PROCESSED_YYYYMMDD/", cas = "umichigan")
Alternatively, each individual dataset can also be provided. For example:
plasma.untargeted.merged <- merge_all_metabolomics(m_m_n = metadata_metabolites_named, m_m_u = metadata_metabolites_unnamed, m_s_n = metadata_sample_named, r_n = results_named, r_u = results_unnamed, phase = "PASS1A-06")
Check the function help for details
Help
Additional details for each function can be found by typing, for example:
?merge_all_metabolomicsNeed extra help? Please, submit an issue here providing as many details as possible.