Plots of WGCNA eigenfeatures for each -ome
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/WGCNA_eigenfeature_plots.Rmd
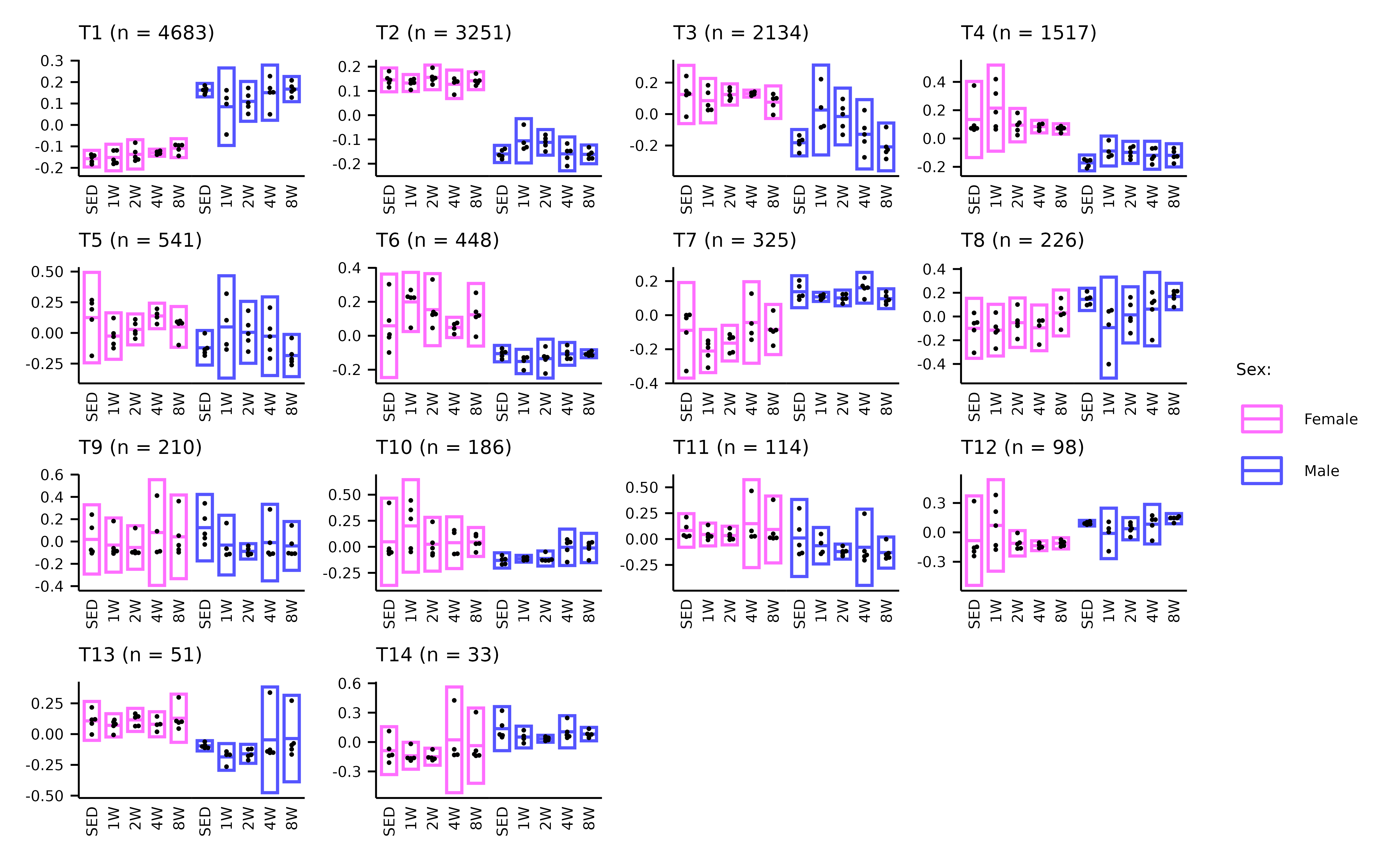
WGCNA_eigenfeature_plots.RmdThis article generates plots of the WGCNA module eigenfeatures (Extended Data Fig. 6A–C).
library(MotrpacRatTraining6moWAT) # plot_eigenfeature
library(MotrpacRatTraining6moWATData)
library(ggplot2)
save_plots <- dir.exists(paths = file.path("..", "..", "plots"))
# Proteomics
p1 <- plot_eigenfeature(x = PROT_WGCNA)
p1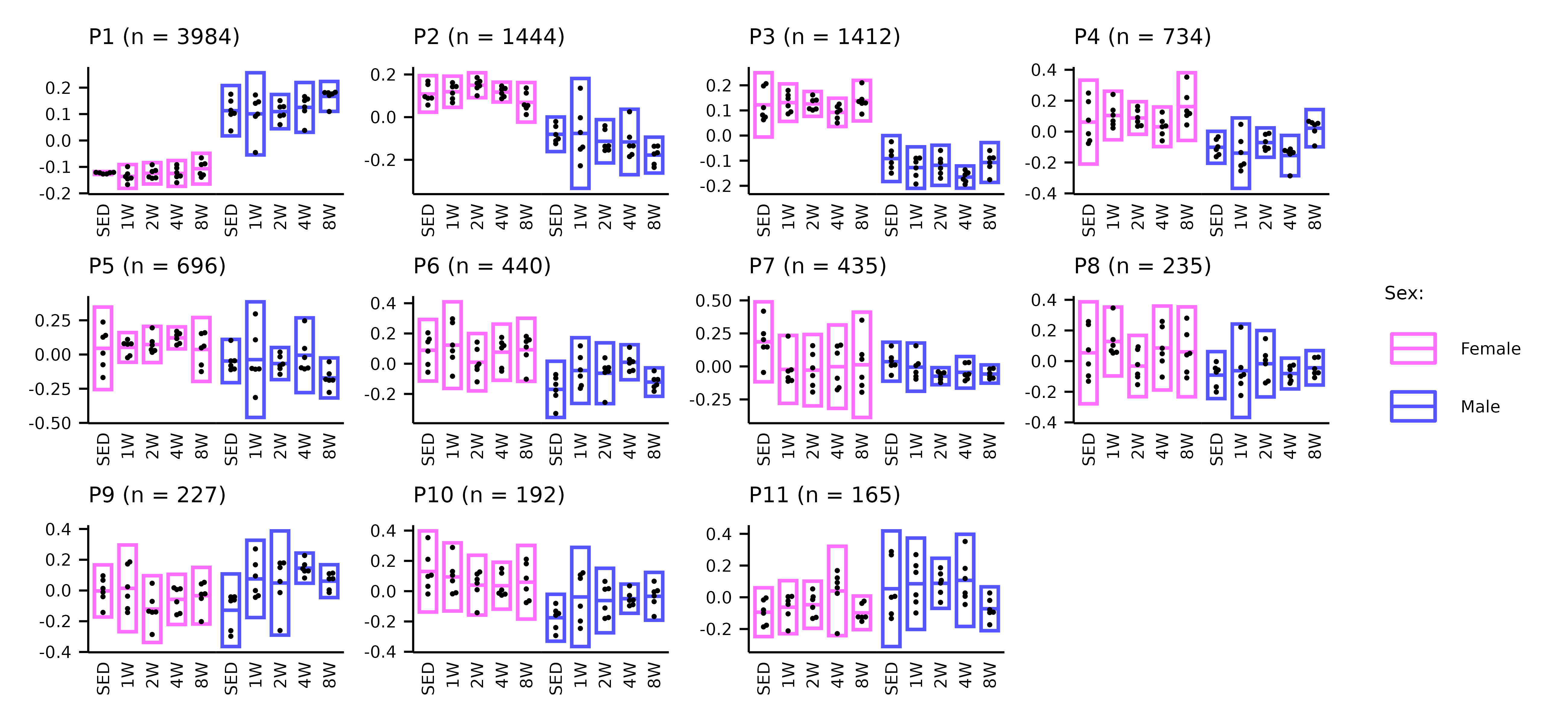
ggsave(file.path("..", "..", "plots", "PROT_module_eigenfeatures.pdf"), p1,
height = 3, width = 6.5, family = "ArialMT")
# Metabolomics
p2 <- plot_eigenfeature(x = METAB_WGCNA)
p2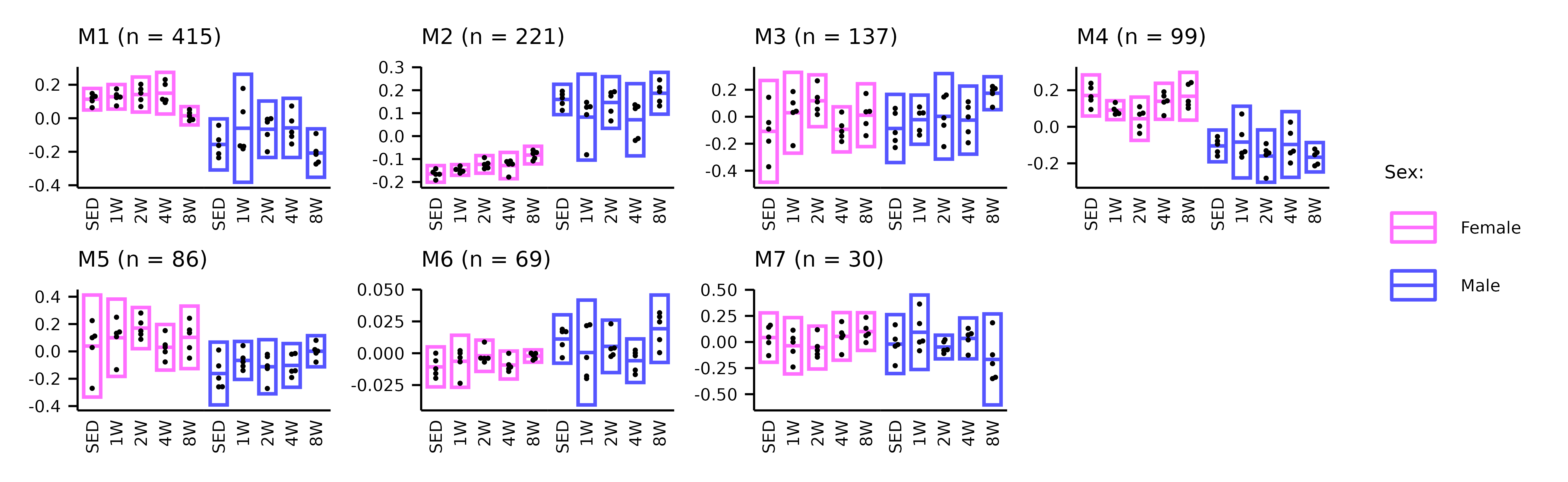
ggsave(file.path("..", "..", "plots", "METAB_module_eigenfeatures.pdf"), p2,
height = 2, width = 6.5, family = "ArialMT")
# Transcriptomics
p3 <- plot_eigenfeature(x = TRNSCRPT_WGCNA)
p3