Plots of log2 relative abundances of various proteins
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/plot_proteins.Rmd
plot_proteins.RmdThis article generates plots of various proteins: Acaca, Fabp4, Lipe, Plin1, and Adipoq (Extended Data Fig. 7D, E). Most were included as plots in the first round of reviewer responses, rather than in the manuscript figures.
library(MotrpacRatTraining6moWATData)
library(MotrpacRatTraining6moData)
library(Biobase)
library(dplyr)
library(tidyr)
library(ggplot2)
library(ggsignif)
library(latex2exp)
library(ggbeeswarm)
genes <- c("Acaca", "Fabp4", "Lipe", "Plin1", "Adipoq")
# Differential analysis results
stats <- PROT_DA$trained_vs_SED %>%
filter(gene_symbol %in% genes,
adj.P.Val < 0.05) %>%
mutate(signif = cut(adj.P.Val,
breaks = c(0, 0.001, 0.01, 0.05, 1),
labels = c("***", "**", "*", ""),
include.lowest = TRUE, right = FALSE,
ordered_result = TRUE),
sex = ifelse(grepl("^F", contrast), "Female", "Male"))
# Individual data points to plot
x <- cbind(select(fData(PROT_EXP), gene_symbol), exprs(PROT_EXP)) %>%
filter(gene_symbol %in% genes) %>%
pivot_longer(cols = -gene_symbol,
names_to = "viallabel",
values_to = "value") %>%
pivot_wider(values_from = value, names_from = gene_symbol) %>%
left_join(pData(PROT_EXP), by = "viallabel")
# Custom plot theme
t1 <- theme_bw() +
theme(text = element_text(size = 6.5, color = "black"),
line = element_line(linewidth = 0.3, color = "black"),
axis.ticks = element_line(linewidth = 0.3, color = "black"),
panel.grid = element_blank(),
panel.border = element_blank(),
axis.ticks.x = element_blank(),
axis.text = element_text(size = 5,
color = "black"),
axis.text.x = element_text(size = 6.5, angle = 90, hjust = 1,
vjust = 0.5),
axis.title = element_text(size = 6.5, margin = margin(),
color = "black"),
axis.line = element_line(linewidth = 0.3),
strip.background = element_blank(),
strip.text = element_text(size = 6.5, color = "black",
margin = margin(b = 5, unit = "pt")),
panel.spacing = unit(-1, "pt"),
plot.title = element_text(size = 9, color = "black",
hjust = 0.5, face = "bold"),
plot.subtitle = element_text(size = 6, color = "black"),
legend.position = "none",
strip.placement = "outside"
)Acaca
## Acaca
p_acaca <- ggplot(x, aes(x = timepoint, y = Acaca)) +
stat_summary(geom = "crossbar",
fun.data = ~ mean_cl_normal(.x),
mapping = aes(color = sex),
fatten = 1, linewidth = 0.4) +
geom_point(shape = 16, size = 0.6,
position = position_beeswarm(cex = 3.5,
dodge.width = 0.4)) +
geom_signif(
data = stats,
aes(y_position = 0.8,
xmin = "SED",
xmax = "8W",
annotations = "*"),
textsize = 3,
vjust = 0.25,
tip_length = 0.02,
color = "black",
size = 0.3,
manual = TRUE
) +
facet_grid(~ sex) +
labs(x = NULL,
y = TeX("log$_2$ relative abundance"),
title = "Acaca") +
scale_color_manual(values = c("#ff6eff", "#5555ff")) +
scale_y_continuous(expand = expansion(mult = 5e-3),
breaks = seq(-1.5, 1, 0.5)) +
coord_cartesian(ylim = c(-1.6, 1), clip = "off") +
t1
p_acaca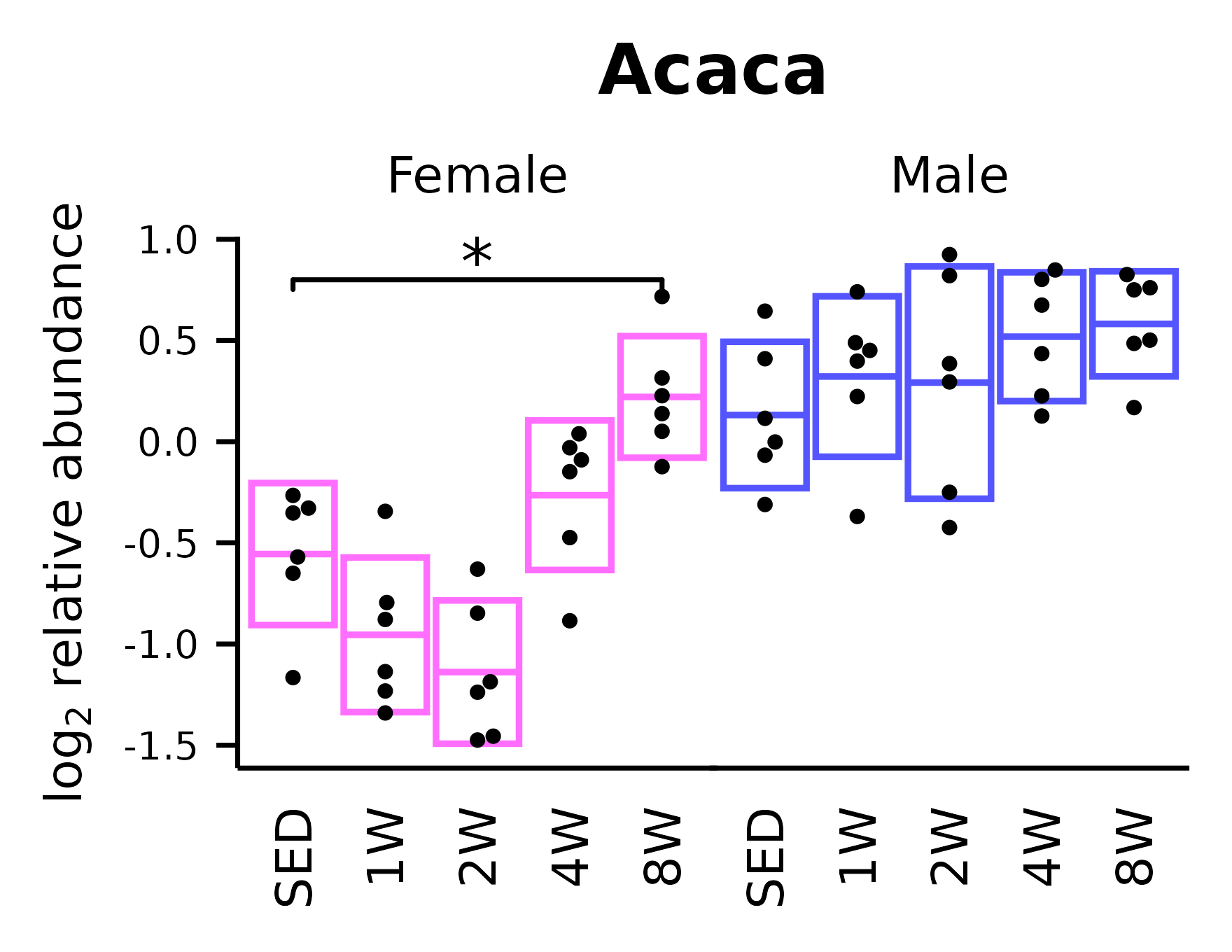
Adipoq
## Adipoq
p_adi <- ggplot(x, aes(x = timepoint, y = Adipoq)) +
stat_summary(geom = "crossbar",
fun.data = ~ mean_cl_normal(.x),
mapping = aes(color = sex),
fatten = 1, linewidth = 0.4) +
geom_point(shape = 16, size = 0.6,
position = position_beeswarm(cex = 3.5,
dodge.width = 0.4)) +
facet_grid(~ sex) +
labs(x = NULL,
y = TeX("log$_2$ relative abundance"),
title = "Adipoq") +
scale_color_manual(values = c("#ff6eff", "#5555ff")) +
scale_y_continuous(expand = expansion(mult = 5e-3),
breaks = seq(-0.4, 0.8, 0.4)) +
coord_cartesian(ylim = c(-0.5, 0.8), clip = "off") +
t1
p_adi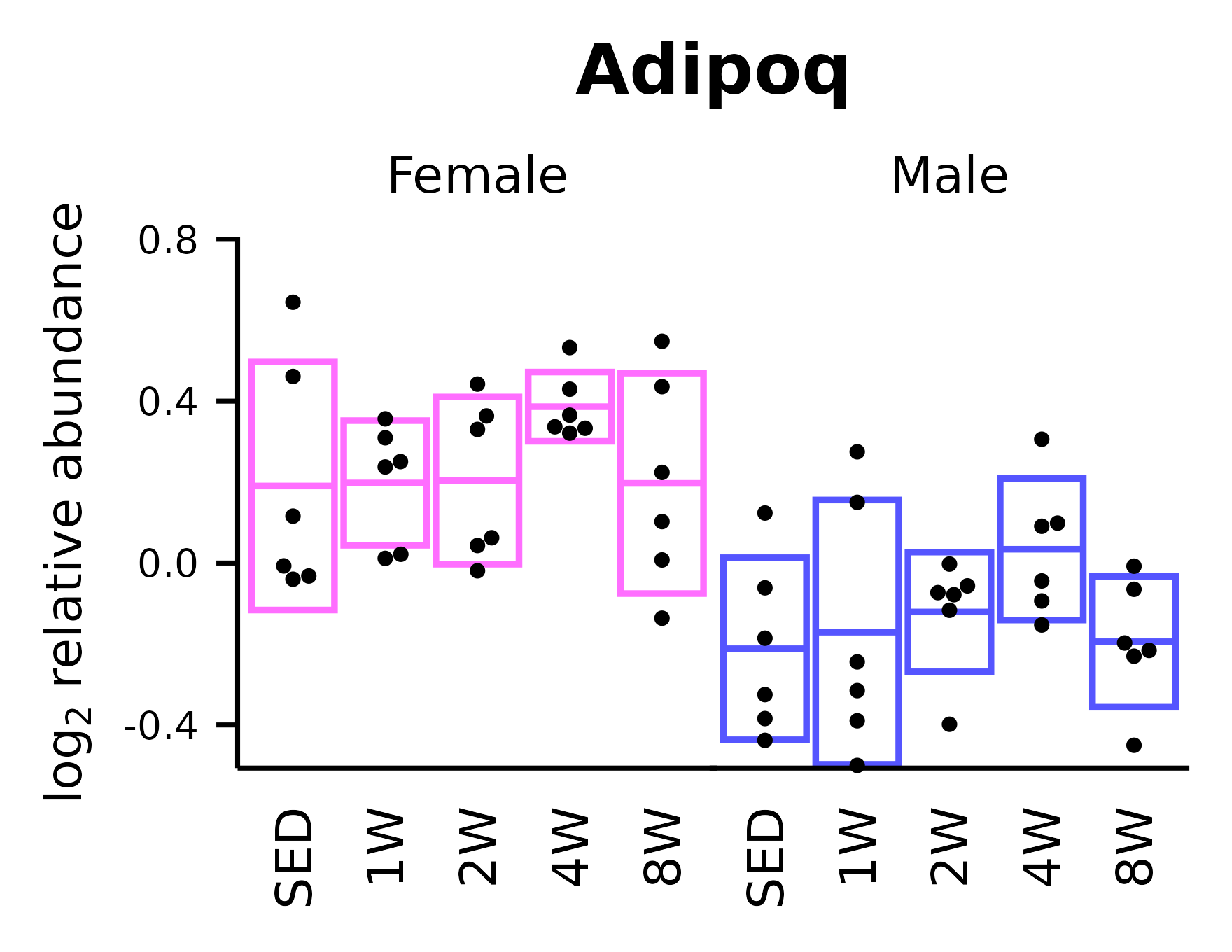
Fabp4
## Fabp4
p_fab <- ggplot(x, aes(x = timepoint, y = Fabp4)) +
stat_summary(geom = "crossbar",
fun.data = ~ mean_cl_normal(.x),
mapping = aes(color = sex),
fatten = 1, linewidth = 0.4) +
geom_point(shape = 16, size = 0.6,
position = position_beeswarm(cex = 3.5,
dodge.width = 0.4)) +
facet_grid(~ sex) +
labs(x = NULL,
y = TeX("log$_2$ relative abundance"),
title = "Fabp4") +
scale_color_manual(values = c("#ff6eff", "#5555ff")) +
scale_y_continuous(expand = expansion(mult = 5e-3),
breaks = seq(-0.8, 0.8, 0.4)) +
coord_cartesian(ylim = c(-0.8, 0.85), clip = "off") +
t1
p_fab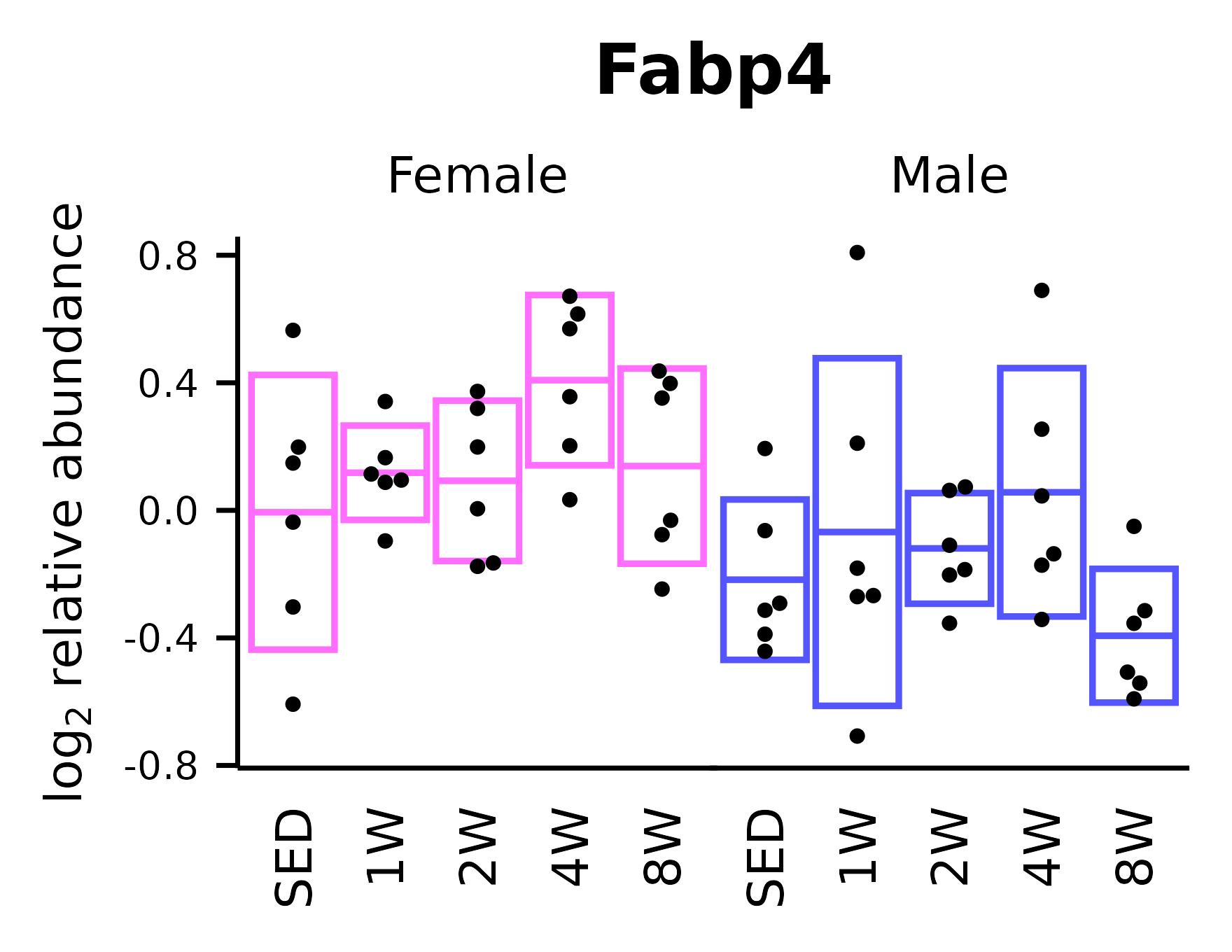
Lipe (Hsl)
## Lipe
p_lipe <- ggplot(x, aes(x = timepoint, y = Lipe)) +
stat_summary(geom = "crossbar",
fun.data = ~ mean_cl_normal(.x),
mapping = aes(color = sex),
fatten = 1, linewidth = 0.4) +
geom_point(shape = 16, size = 0.6,
position = position_beeswarm(cex = 3.5,
dodge.width = 0.4)) +
facet_grid(~ sex) +
labs(x = NULL,
y = TeX("log$_2$ relative abundance"),
title = "Lipe (Hsl)") +
scale_color_manual(values = c("#ff6eff", "#5555ff")) +
scale_y_continuous(expand = expansion(mult = 5e-3),
breaks = seq(-0.8, 0.8, 0.4)) +
coord_cartesian(ylim = c(-0.8, 0.8), clip = "off") +
t1
p_lipe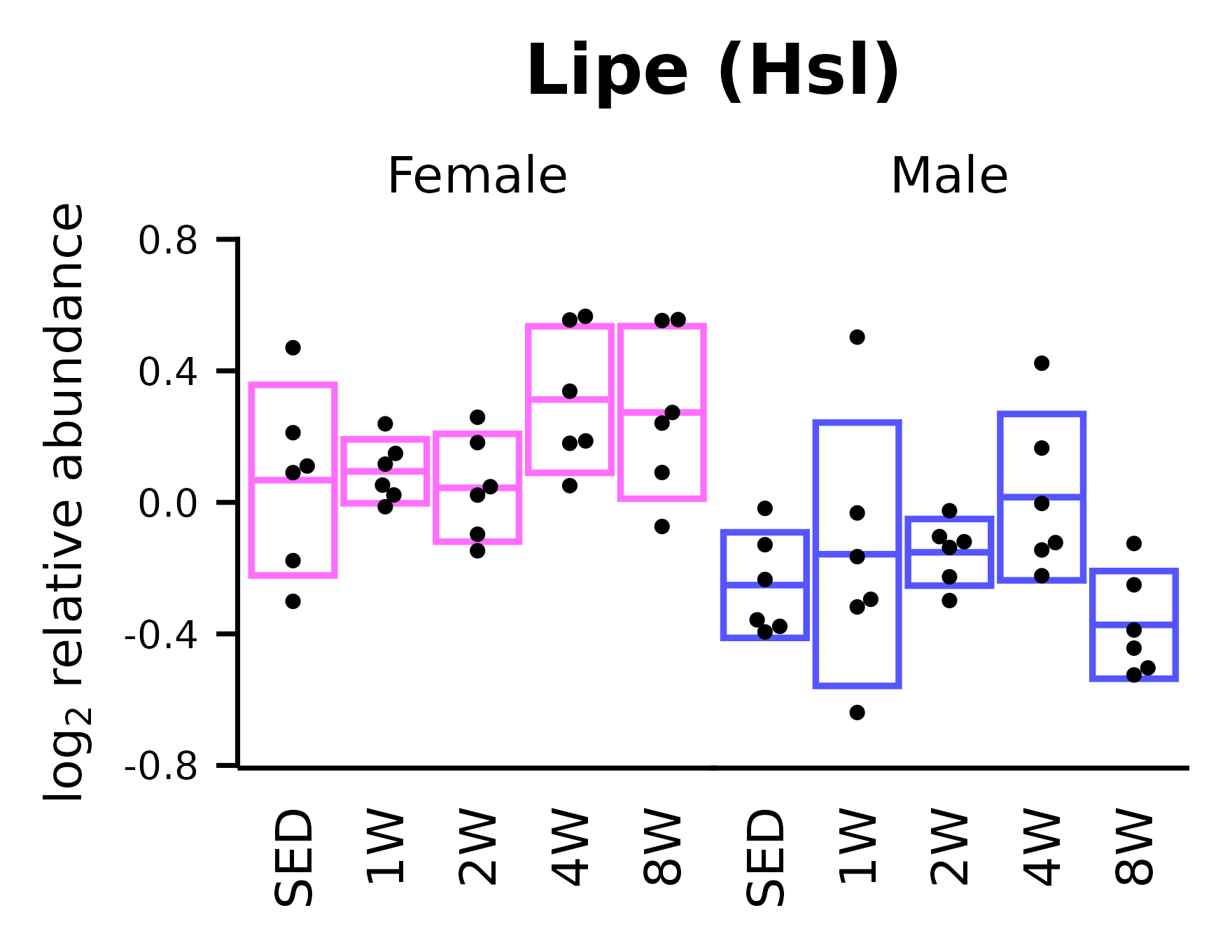
Plin1
## Plin1
p_plin <- ggplot(x, aes(x = timepoint, y = Plin1)) +
stat_summary(geom = "crossbar",
fun.data = ~ mean_cl_normal(.x),
mapping = aes(color = sex),
fatten = 1, linewidth = 0.4) +
geom_point(shape = 16, size = 0.6,
position = position_beeswarm(cex = 3.5,
dodge.width = 0.4)) +
facet_grid(~ sex) +
labs(x = NULL,
y = TeX("log$_2$ relative abundance"),
title = "Plin1") +
scale_color_manual(values = c("#ff6eff", "#5555ff")) +
scale_y_continuous(expand = expansion(mult = 5e-3),
breaks = seq(-0.8, 0.8, 0.4)) +
coord_cartesian(ylim = c(-0.85, 0.8), clip = "off") +
t1
p_plin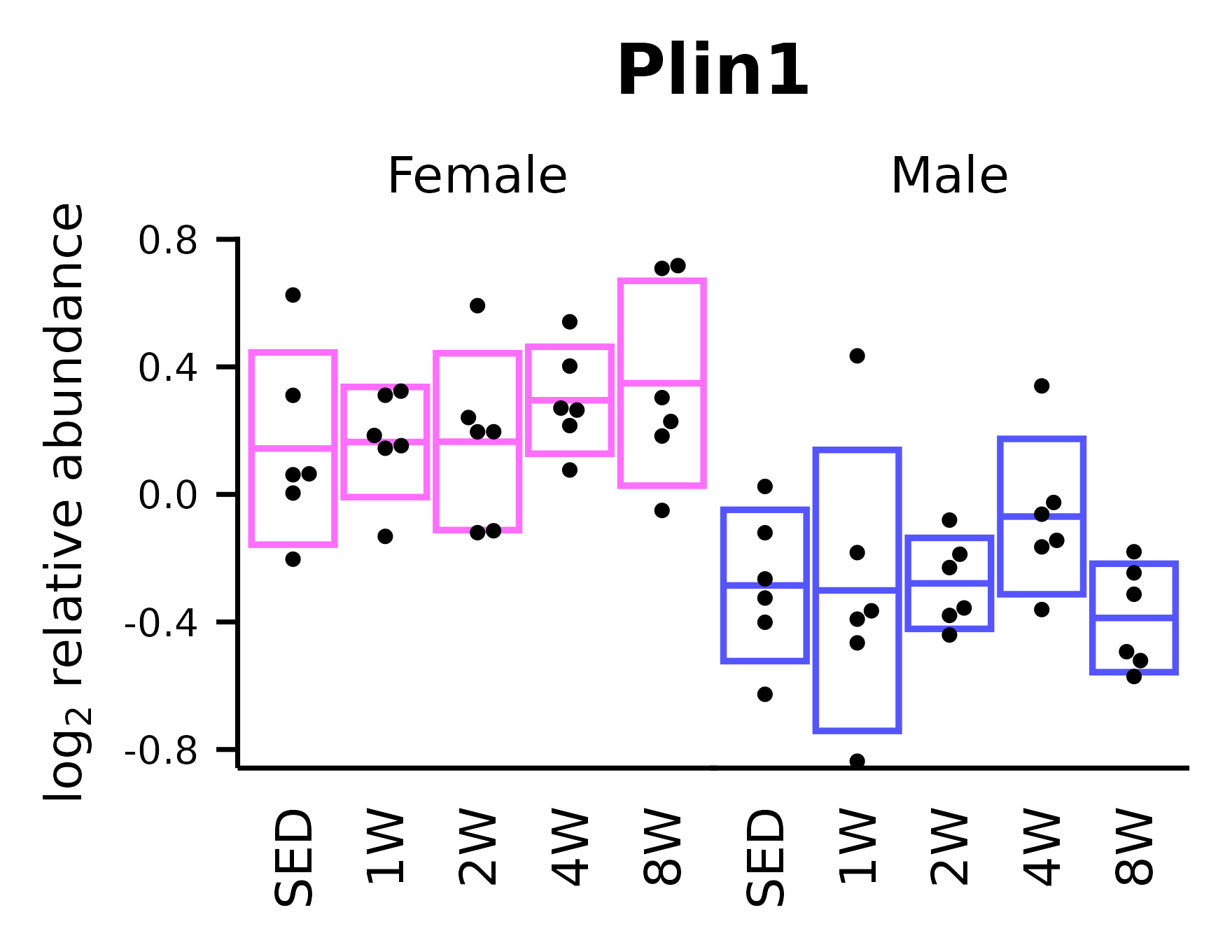
Session Info
sessionInfo()
#> R version 4.4.0 (2024-04-24)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggbeeswarm_0.7.2 latex2exp_0.9.6
#> [3] ggsignif_0.6.4 ggplot2_3.5.1
#> [5] tidyr_1.3.1 dplyr_1.1.4
#> [7] Biobase_2.64.0 BiocGenerics_0.50.0
#> [9] MotrpacRatTraining6moData_2.0.0 MotrpacRatTraining6moWATData_2.0.0
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.5 beeswarm_0.4.0 xfun_0.43 bslib_0.7.0
#> [5] htmlwidgets_1.6.4 vctrs_0.6.5 tools_4.4.0 generics_0.1.3
#> [9] tibble_3.2.1 fansi_1.0.6 highr_0.10 cluster_2.1.6
#> [13] pkgconfig_2.0.3 data.table_1.15.4 checkmate_2.3.1 desc_1.4.3
#> [17] lifecycle_1.0.4 compiler_4.4.0 farver_2.1.1 stringr_1.5.1
#> [21] textshaping_0.3.7 munsell_0.5.1 vipor_0.4.7 htmltools_0.5.8.1
#> [25] sass_0.4.9 yaml_2.3.8 htmlTable_2.4.2 Formula_1.2-5
#> [29] pillar_1.9.0 pkgdown_2.0.9 jquerylib_0.1.4 cachem_1.0.8
#> [33] Hmisc_5.1-2 rpart_4.1.23 tidyselect_1.2.1 digest_0.6.35
#> [37] stringi_1.8.3 purrr_1.0.2 fastmap_1.1.1 grid_4.4.0
#> [41] colorspace_2.1-0 cli_3.6.2 magrittr_2.0.3 base64enc_0.1-3
#> [45] utf8_1.2.4 foreign_0.8-86 withr_3.0.0 scales_1.3.0
#> [49] backports_1.4.1 rmarkdown_2.26 nnet_7.3-19 gridExtra_2.3
#> [53] ragg_1.3.0 memoise_2.0.1 evaluate_0.23 knitr_1.46
#> [57] rlang_1.1.3 glue_1.7.0 rstudioapi_0.16.0 jsonlite_1.8.8
#> [61] R6_2.5.1 systemfonts_1.0.6 fs_1.6.4