Plots of plasma clinical analytes
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/plot_analytes.Rmd
plot_analytes.RmdThis article generates plots of the plasma clinical analytes (Fig. 1F–I, Extended Data Fig. 1E, F).
# Required packages
library(MotrpacRatTraining6moWAT) # plot_baseline
library(MotrpacRatTraining6moWATData)
library(ggplot2)
library(dplyr)
library(purrr)
library(tidyr)
library(tibble)
library(emmeans)
library(scales)
save_plots <- dir.exists(paths = file.path("..", "..", "plots"))
x <- ANALYTES %>%
filter(omics_subset) %>%
dplyr::rename(group = timepoint)
stats_df <- ANALYTES_STATS$timewise
# Reformat confidence interval data
conf_df <- map(ANALYTES_EMM$timewise, function(emm_i) {
terms_i <- attr(terms(emm_i@model.info), which = "term.labels")
out <- summary(emm_i) %>%
as.data.frame() %>%
dplyr::rename(any_of(c(lower.CL = "asymp.LCL",
upper.CL = "asymp.UCL",
response_mean = "response",
response_mean = "rate")))
out <- out %>%
mutate(timepoint = factor(timepoint,
levels = c("SED", paste0(2 ^ (0:3), "W"))))
return(out)
}) %>%
enframe(name = "response") %>%
unnest(value) %>%
dplyr::rename(group = timepoint)Glucagon
## Glucagon
plot_baseline(x, response = "glucagon",
conf = filter(conf_df, response == "Glucagon"),
stats = filter(stats_df, response == "Glucagon"),
bracket.nudge.y = 12) +
scale_y_continuous(name = "Glucagon (pM)",
breaks = seq(0, 160, 40),
expand = expansion(mult = 5e-3)) +
coord_cartesian(ylim = c(0, 160), clip = "off") +
theme(plot.margin = margin(t = 4, r = 4, b = 4, l = 4),
strip.text = element_text(margin = margin(b = 14)))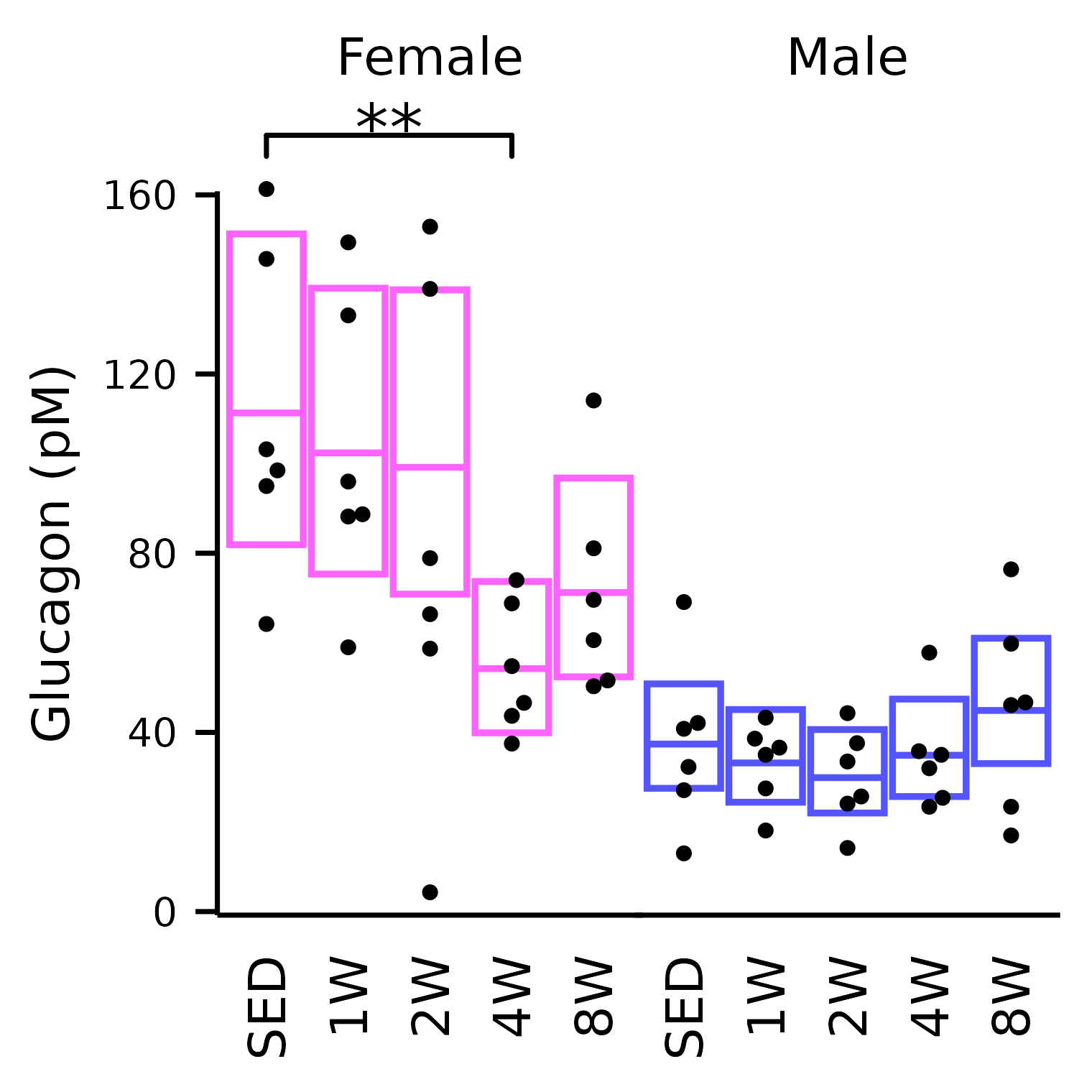
Glucose
## Glucose
plot_baseline(x, response = "glucose",
conf = filter(conf_df, response == "Glucose"),
stats = filter(stats_df, response == "Glucose"),
bracket.nudge.y = 6) +
scale_y_continuous(name = "Glucose (mg/dL)",
expand = expansion(mult = 5e-3)) +
coord_cartesian(ylim = c(115, 200), clip = "off") +
theme(plot.margin = margin(t = 4, r = 4, b = 4, l = 4),
strip.text = element_text(margin = margin(b = 4)))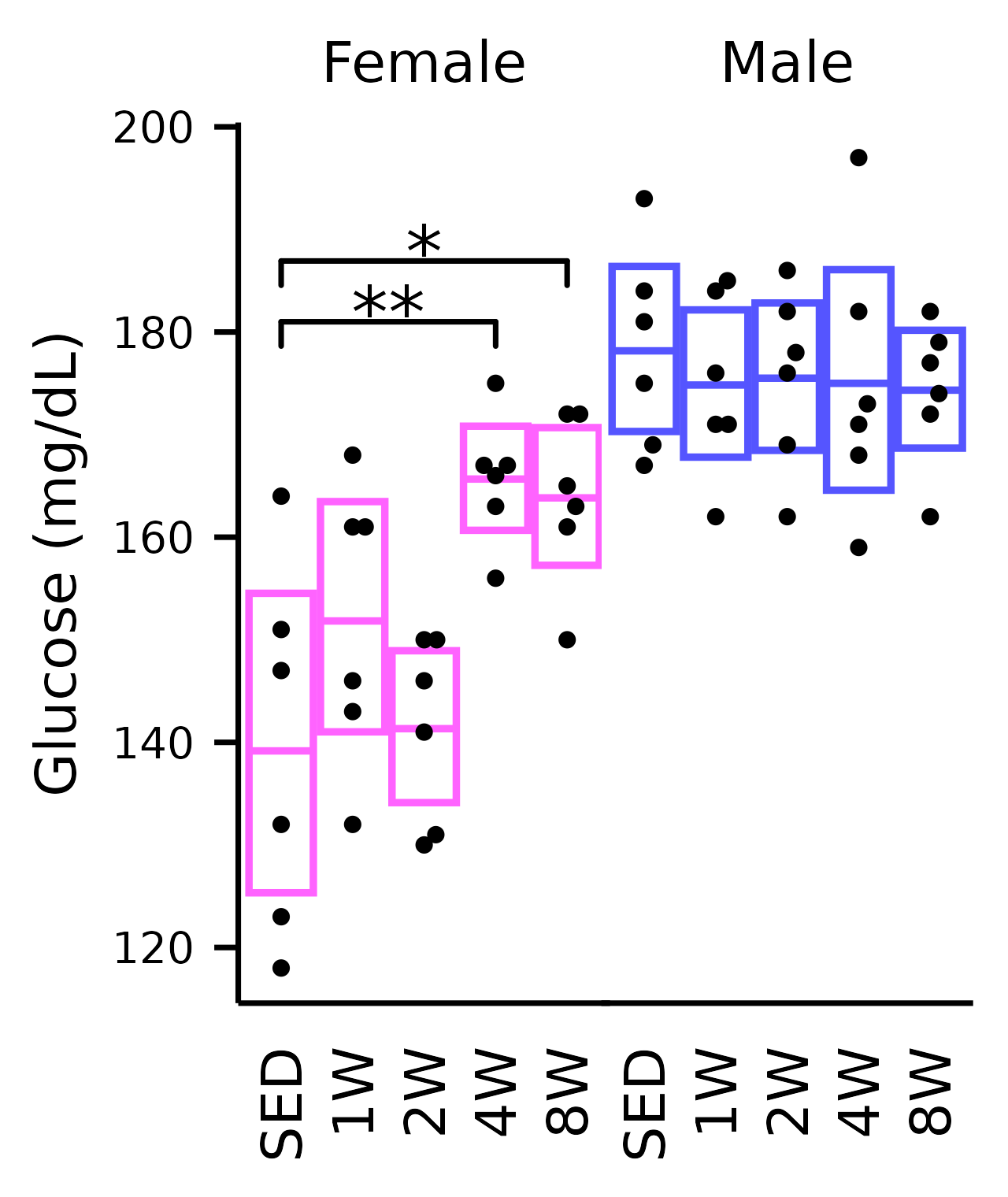
Glycerol
## Glycerol
plot_baseline(x, response = "glycerol",
conf = filter(conf_df, response == "Glycerol"),
stats = filter(stats_df, response == "Glycerol"),
bracket.nudge.y = 0.2) +
scale_y_continuous(name = "Glycerol (mg/dL)",
expand = expansion(mult = 5e-3)) +
coord_cartesian(ylim = c(0, 3), clip = "off") +
theme(plot.margin = margin(t = 4, r = 4, b = 4, l = 4),
strip.text = element_text(margin = margin(b = 5)))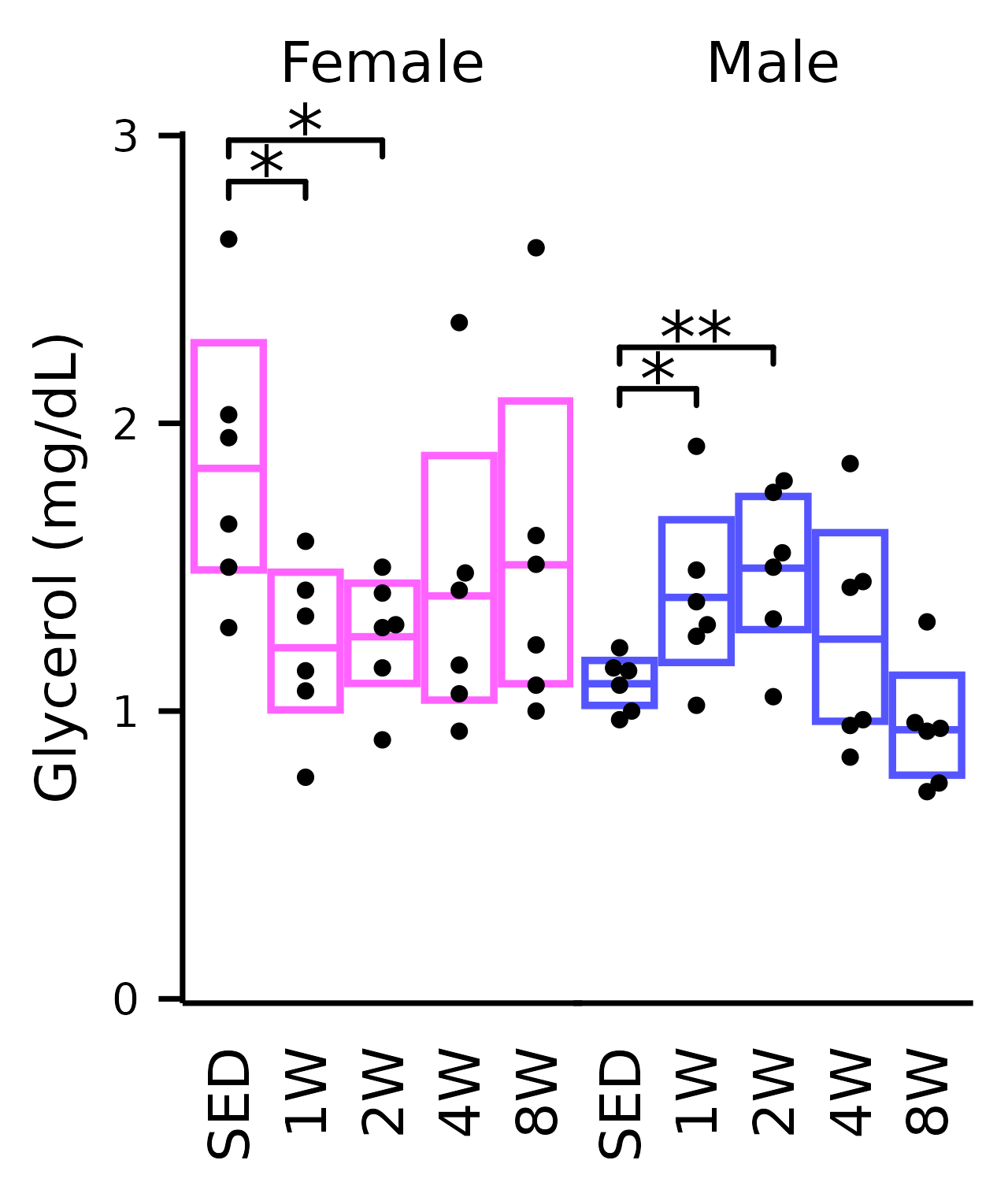
Insulin
## Insulin
plot_baseline(x, response = "insulin_iu",
bracket.nudge.y = 10) +
scale_y_continuous(name = "Insulin (mg/dL)",
expand = expansion(mult = 5e-3),
breaks = seq(20, 140, 20),
sec.axis = sec_axis(name = "Insulin (pg/mL)",
trans = ~ .x / 0.023,
breaks = 1000 * 1:6)) +
coord_cartesian(ylim = c(20, 140), clip = "off") +
theme(plot.margin = margin(t = 4, r = 4, b = 4, l = 4),
strip.text = element_text(margin = margin(b = 5)))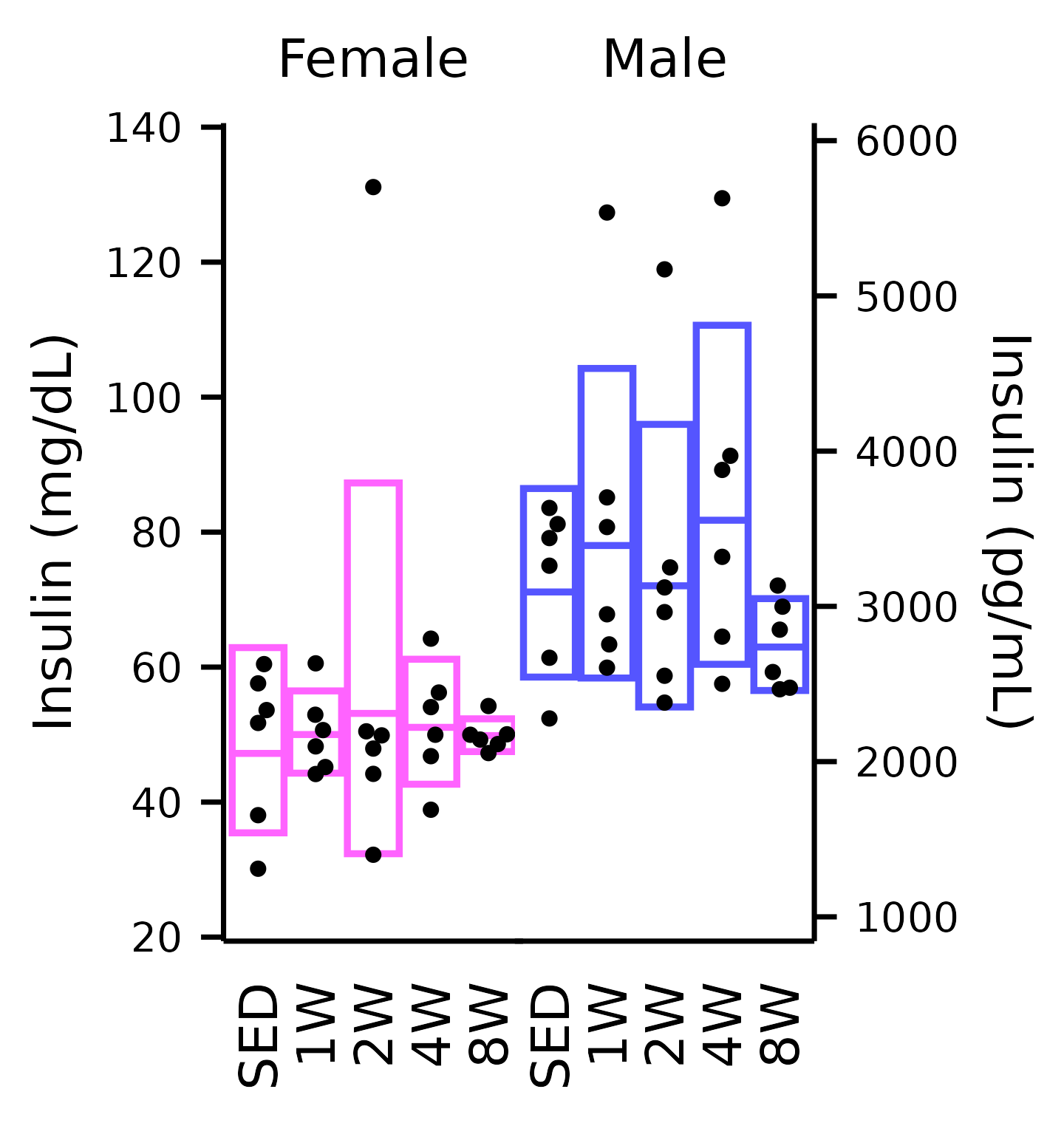
Leptin
## Leptin
plot_baseline(x, response = "leptin",
conf = filter(conf_df, response == "Leptin"),
stats = filter(stats_df, response == "Leptin"),
bracket.nudge.y = 1e4 / 3) +
scale_y_continuous(name = "Leptin (pg/mL)",
labels = scales::label_scientific(digits = 1),
expand = expansion(mult = 5e-3)) +
coord_cartesian(ylim = c(0, 6e4), clip = "off") +
theme(plot.margin = margin(t = 4, r = 4, b = 4, l = 4),
strip.text = element_text(margin = margin(b = 14)))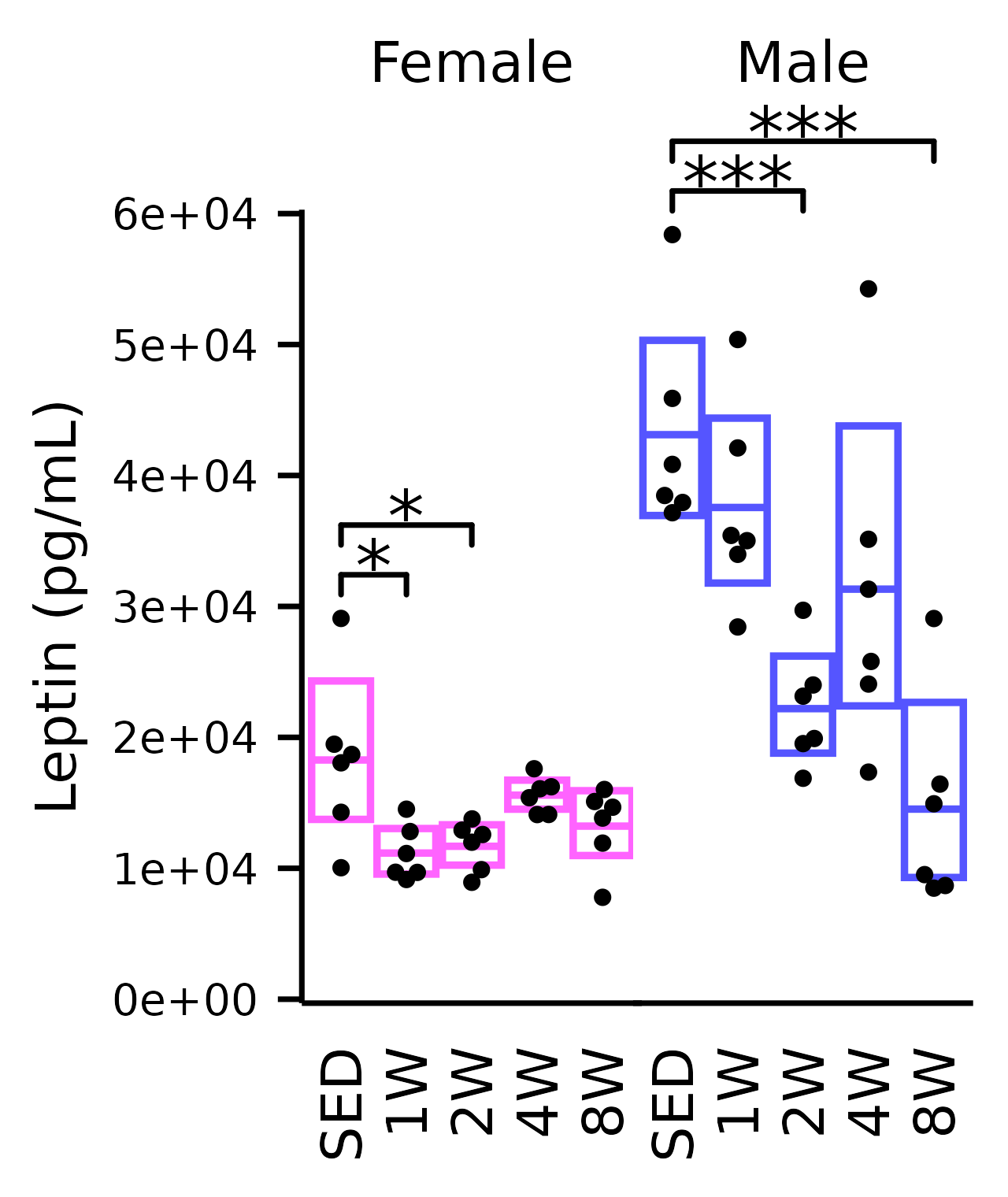
NEFA
## NEFA
plot_baseline(x, response = "nefa",
conf = filter(conf_df, response == "NEFA"),
stats = filter(stats_df, response == "NEFA"),
bracket.nudge.y = 0.1) +
scale_y_continuous(name = "NEFA (mmol/L)",
breaks = seq(0.3, 1.3, 0.2),
expand = expansion(mult = 5e-3)) +
coord_cartesian(ylim = c(0.3, 1.3), clip = "off") +
theme(plot.margin = margin(t = 4, r = 4, b = 4, l = 4),
strip.text = element_text(margin = margin(b = 10)))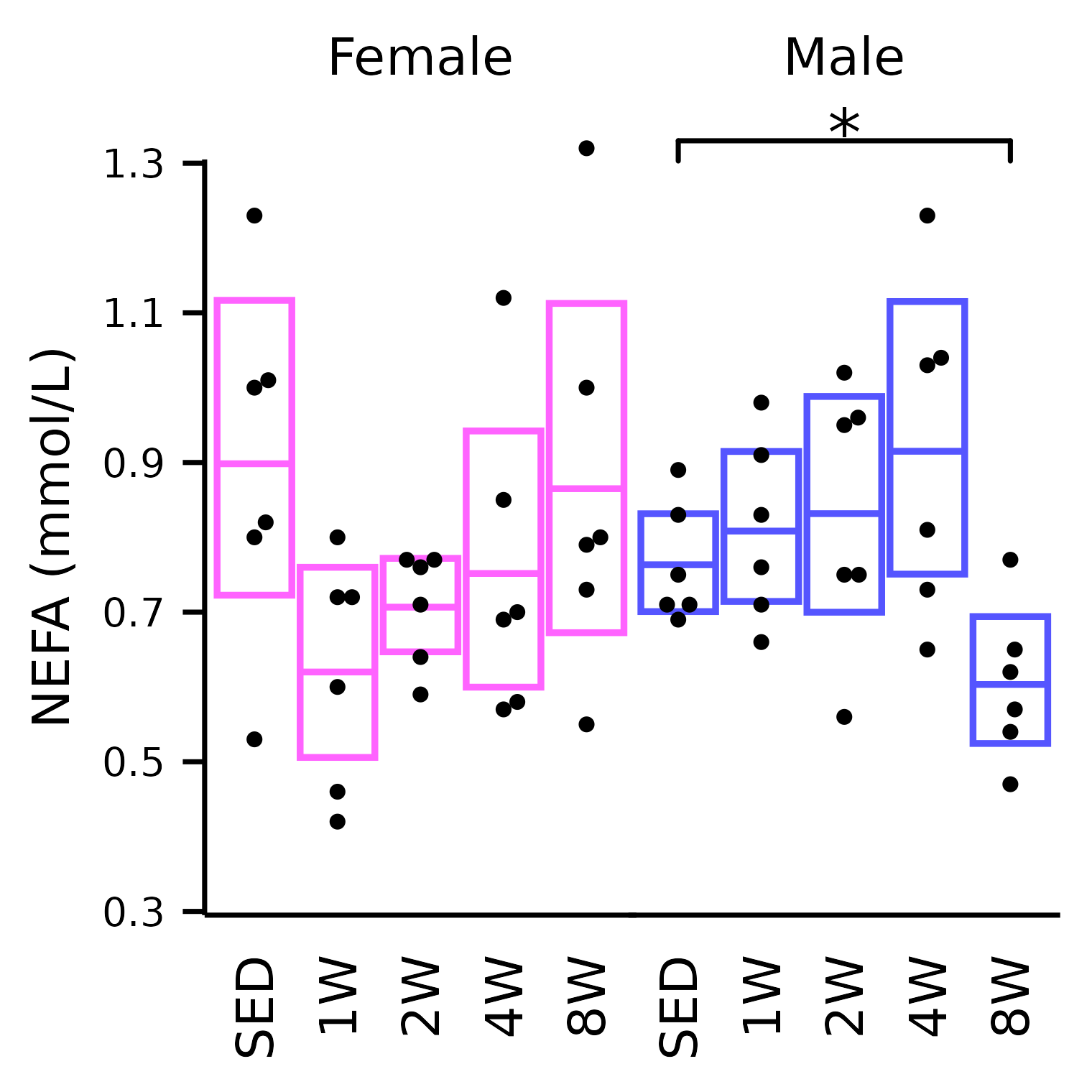
Session Info
sessionInfo()
#> R version 4.4.0 (2024-04-24)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] scales_1.3.0 emmeans_1.10.1
#> [3] tibble_3.2.1 tidyr_1.3.1
#> [5] purrr_1.0.2 dplyr_1.1.4
#> [7] ggplot2_3.5.1 MotrpacRatTraining6moWATData_2.0.0
#> [9] MotrpacRatTraining6moWAT_1.0.1 Biobase_2.64.0
#> [11] BiocGenerics_0.50.0
#>
#> loaded via a namespace (and not attached):
#> [1] RColorBrewer_1.1-3 rstudioapi_0.16.0 jsonlite_1.8.8
#> [4] shape_1.4.6.1 magrittr_2.0.3 estimability_1.5
#> [7] ggbeeswarm_0.7.2 farver_2.1.1 rmarkdown_2.26
#> [10] GlobalOptions_0.1.2 fs_1.6.4 zlibbioc_1.50.0
#> [13] ragg_1.3.0 vctrs_0.6.5 memoise_2.0.1
#> [16] base64enc_0.1-3 rstatix_0.7.2 htmltools_0.5.8.1
#> [19] dynamicTreeCut_1.63-1 curl_5.2.1 broom_1.0.5
#> [22] Formula_1.2-5 sass_0.4.9 bslib_0.7.0
#> [25] htmlwidgets_1.6.4 desc_1.4.3 impute_1.78.0
#> [28] cachem_1.0.8 lifecycle_1.0.4 iterators_1.0.14
#> [31] pkgconfig_2.0.3 Matrix_1.7-0 R6_2.5.1
#> [34] fastmap_1.1.1 GenomeInfoDbData_1.2.12 clue_0.3-65
#> [37] digest_0.6.35 colorspace_2.1-0 patchwork_1.2.0
#> [40] AnnotationDbi_1.65.2 S4Vectors_0.42.0 textshaping_0.3.7
#> [43] Hmisc_5.1-2 RSQLite_2.3.6 ggpubr_0.6.0
#> [46] labeling_0.4.3 filelock_1.0.3 latex2exp_0.9.6
#> [49] fansi_1.0.6 httr_1.4.7 abind_1.4-5
#> [52] compiler_4.4.0 withr_3.0.0 bit64_4.0.5
#> [55] doParallel_1.0.17 htmlTable_2.4.2 backports_1.4.1
#> [58] BiocParallel_1.38.0 carData_3.0-5 DBI_1.2.2
#> [61] highr_0.10 ggsignif_0.6.4 rjson_0.2.21
#> [64] tools_4.4.0 vipor_0.4.7 foreign_0.8-86
#> [67] beeswarm_0.4.0 msigdbr_7.5.1 nnet_7.3-19
#> [70] glue_1.7.0 grid_4.4.0 checkmate_2.3.1
#> [73] cluster_2.1.6 fgsea_1.30.0 generics_0.1.3
#> [76] gtable_0.3.5 preprocessCore_1.66.0 data.table_1.15.4
#> [79] WGCNA_1.72-5 car_3.1-2 utf8_1.2.4
#> [82] XVector_0.44.0 foreach_1.5.2 pillar_1.9.0
#> [85] stringr_1.5.1 babelgene_22.9 limma_3.60.0
#> [88] circlize_0.4.16 splines_4.4.0 BiocFileCache_2.12.0
#> [91] lattice_0.22-6 survival_3.5-8 bit_4.0.5
#> [94] tidyselect_1.2.1 GO.db_3.19.1 ComplexHeatmap_2.20.0
#> [97] locfit_1.5-9.9 Biostrings_2.72.0 knitr_1.46
#> [100] gridExtra_2.3 IRanges_2.38.0 edgeR_4.2.0
#> [103] stats4_4.4.0 xfun_0.43 statmod_1.5.0
#> [106] matrixStats_1.3.0 stringi_1.8.3 UCSC.utils_1.0.0
#> [109] yaml_2.3.8 evaluate_0.23 codetools_0.2-20
#> [112] cli_3.6.2 ontologyIndex_2.12 rpart_4.1.23
#> [115] systemfonts_1.0.6 munsell_0.5.1 jquerylib_0.1.4
#> [118] Rcpp_1.0.12 GenomeInfoDb_1.40.0 dbplyr_2.5.0
#> [121] png_0.1-8 fastcluster_1.2.6 parallel_4.4.0
#> [124] pkgdown_2.0.9 blob_1.2.4 mvtnorm_1.2-4
#> [127] crayon_1.5.2 GetoptLong_1.0.5 rlang_1.1.3
#> [130] cowplot_1.1.3 fastmatch_1.1-4 KEGGREST_1.44.0