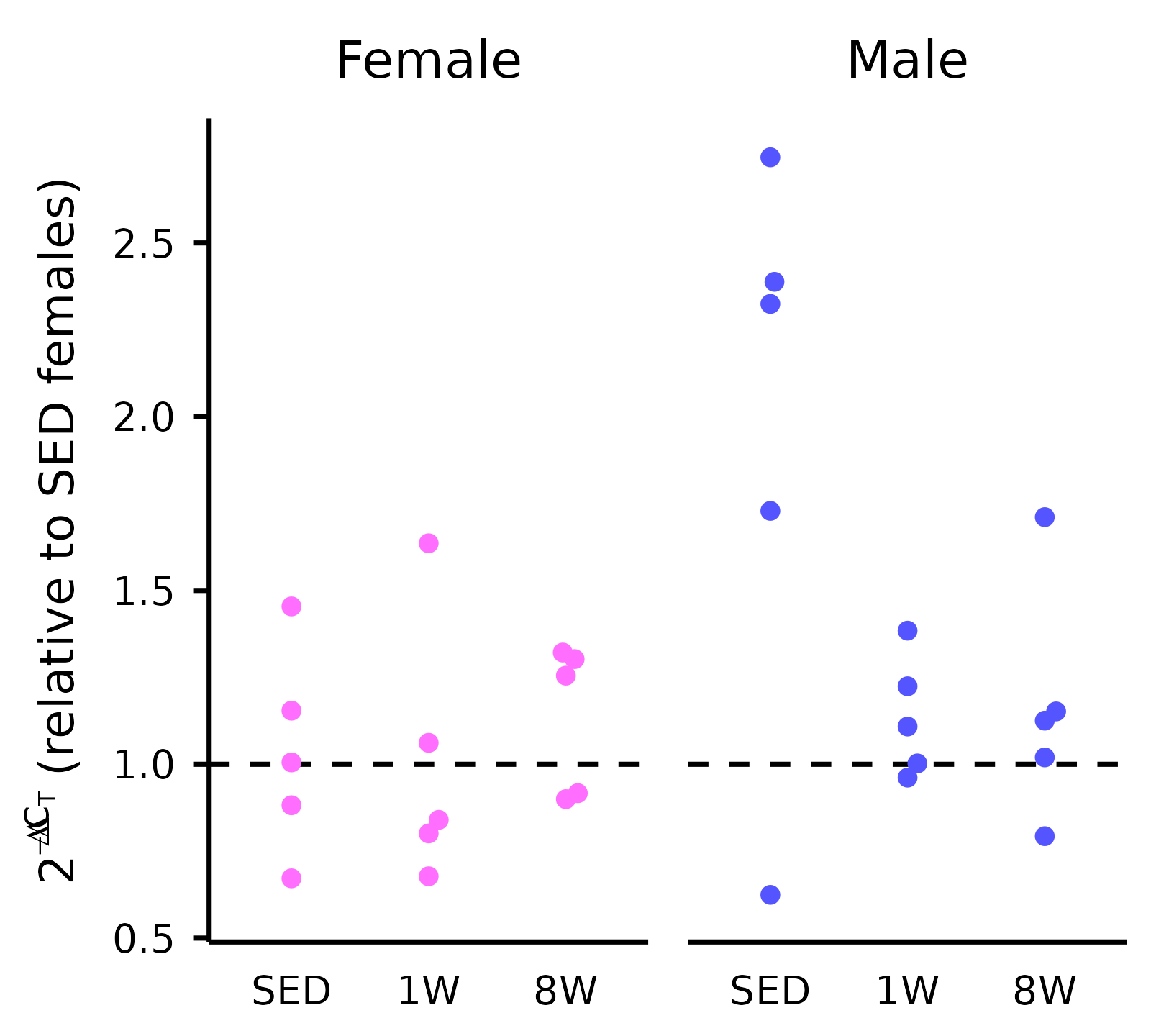
Plot of Mitochondrial DNA (mtDNA)
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/plot_mtDNA.Rmd
plot_mtDNA.RmdThis article generates a plot of the mitochondrial DNA (Extended Data Fig. 7A).
# Required packages
library(MotrpacRatTraining6moWATData)
library(MotrpacRatTraining6moWAT)
library(ggplot2)
library(ggbeeswarm)
library(latex2exp)
p1 <- ggplot(MITO_DNA, aes(x = timepoint, y = relative_expr)) +
facet_wrap(~ sex, drop = FALSE, nrow = 1) +
geom_hline(yintercept = 1, lty = "dashed",
color = "black", linewidth = 0.3) +
ggbeeswarm::geom_beeswarm(aes(color = sex),
cex = 3, size = 0.5) +
# ggpubr::stat_compare_means(method = "kruskal",
# label.sep = ":\n",
# label.x = 1.5, label.y = 2.3,
# hjust = 0, vjust = 1, size = 1.76) +
scale_y_continuous(limits = c(0.5, NA),
breaks = seq(0.5, 2.5, 0.5),
expand = expansion(mult = c(5e-3, 0.05))) +
scale_color_manual(values = c("#ff6eff", "#5555ff")) +
guides(color = guide_none()) +
labs(x = NULL,
y = TeX("$2^{- \\Delta \\Delta C_T}$ (relative to SED females)")) +
theme_pub() +
theme(strip.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.length = unit(2, "pt"),
plot.margin = unit(c(0, rep(3, 3)), "pt"))
p1