Plots of plasma clinical analytes
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/analyte_plots.Rmd
analyte_plots.Rmd
# Required packages
library(MotrpacRatTrainingPhysiologyData)
library(dplyr)
library(tibble)
library(tidyr)
library(purrr)
library(ggplot2)
library(latex2exp)Overview
This article generates all plasma clinical analyte figures. A bracket
indicates a significant difference (Dunnett’s test p-value < 0.05)
between the means of the SED group and a trained group (1W, 2W, 4W, 8W).
Plots are generated for each combination of age and sex and such that
the y-axis limits for the same analyte are identical. This allows for
easier visual comparisons. Please refer to the “Statistical analyses of
plasma clinical analytes” vignette for code to generate
ANALYTES_STATS.
Data Preparation:
We apply the summary method to each emmGrid
object and rename columns to work with plot_baseline. Since
the formula for corticosterone did not include age as a
predictor, the entries for the age column are
NA. We instead modify these entries to include both levels
of age separated by a comma. Using this, we can separate
the rows to effectively duplicate the results so that we now have
corticosterone data for both ages. Lastly, we ensure that
group is a factor to preserve the order when plotting. This
list of objects is then converted to a single data.frame
with the response column specifying which analyte is being
tested.
# Reformat confidence interval data
conf_df <- map(ANALYTES_EMM, function(emm_i) {
terms_i <- attr(terms(emm_i@model.info), which = "term.labels")
out <- summary(emm_i) %>%
as.data.frame() %>%
rename(any_of(c(lower.CL = "asymp.LCL",
upper.CL = "asymp.UCL",
response_mean = "response",
response_mean = "rate")))
# In some cases, age or sex is not included in the model, so their entries
# would be NA. We will instead duplicate those rows for plotting.
# This was already done for ANALYTES_STATS.
if (!"age" %in% colnames(out)) {
out$age <- "6M, 18M"
}
out <- out %>%
separate_longer_delim(cols = age, delim = ", ") %>%
mutate(group = factor(group, levels = c("SED", paste0(2 ^ (0:3), "W"))))
return(out)
}) %>%
enframe(name = "response") %>%
unnest(value)Corticosterone
Corticosterone (ng/mL).
6M Female
plot_baseline(ANALYTES,
response = "corticosterone",
conf = filter(conf_df, response == "Corticosterone"),
stats = filter(ANALYTES_STATS, response == "Corticosterone"),
sex = "Female", age = "6M") +
scale_y_continuous(name = "Corticosterone (ng/mL)",
breaks = seq(0, 170, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 170), clip = "off") +
theme(plot.margin = unit(c(20, 5, 5, 5), units = "pt"))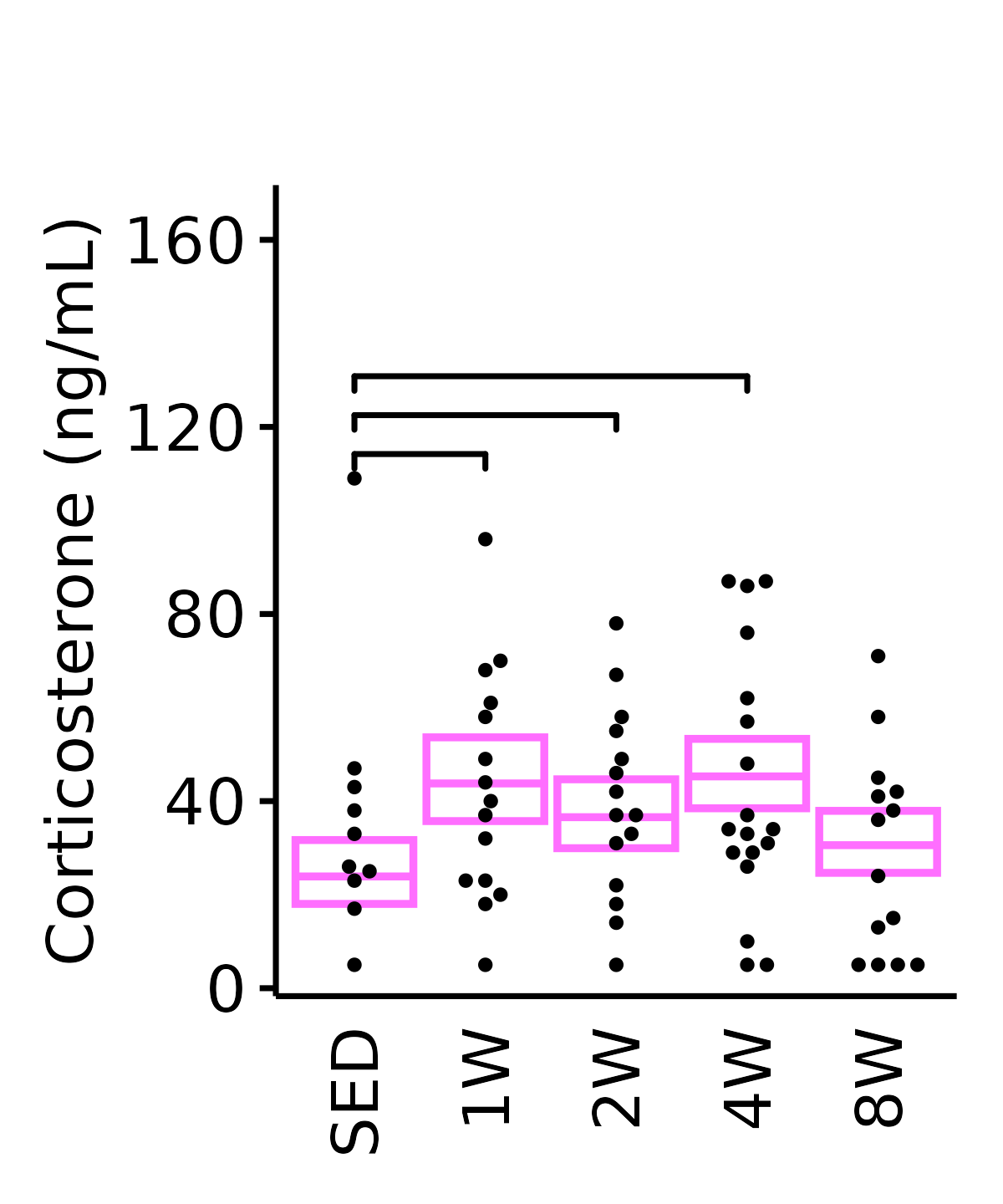
6M Male
plot_baseline(ANALYTES,
response = "corticosterone",
conf = filter(conf_df, response == "Corticosterone"),
stats = filter(ANALYTES_STATS,
response == "Corticosterone"),
sex = "Male", age = "6M", y_position = 145) +
scale_y_continuous(name = "Corticosterone (ng/mL)",
breaks = seq(0, 170, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 170), clip = "off") +
theme(plot.margin = unit(c(20, 5, 5, 5), units = "pt"))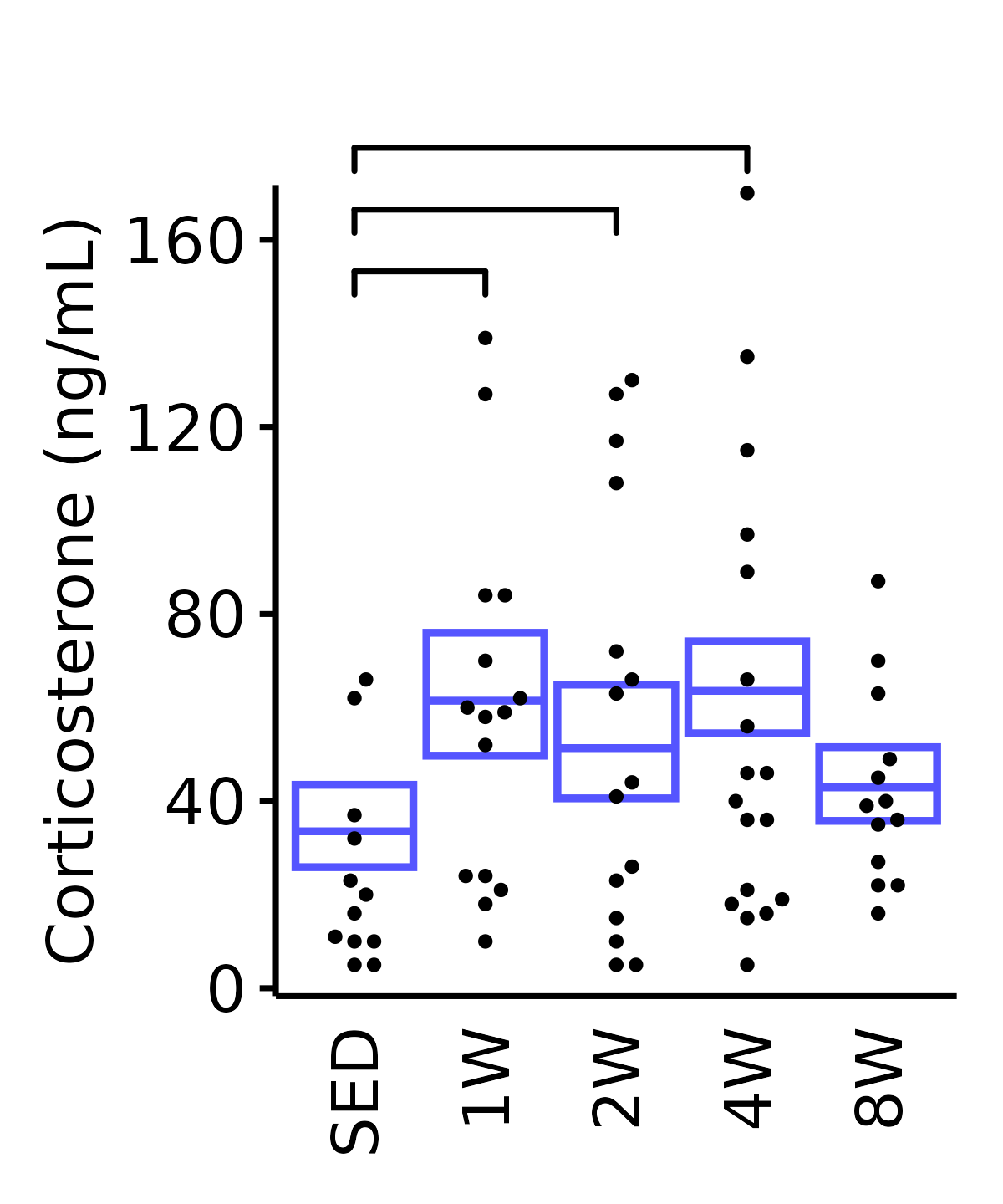
18M Female
plot_baseline(ANALYTES,
response = "corticosterone",
conf = filter(conf_df, response == "Corticosterone"),
stats = filter(ANALYTES_STATS,
response == "Corticosterone"),
sex = "Female", age = "18M", y_position = 75) +
scale_y_continuous(name = "Corticosterone (ng/mL)",
breaks = seq(0, 170, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 170), clip = "off") +
theme(plot.margin = unit(c(20, 5, 5, 5), units = "pt"))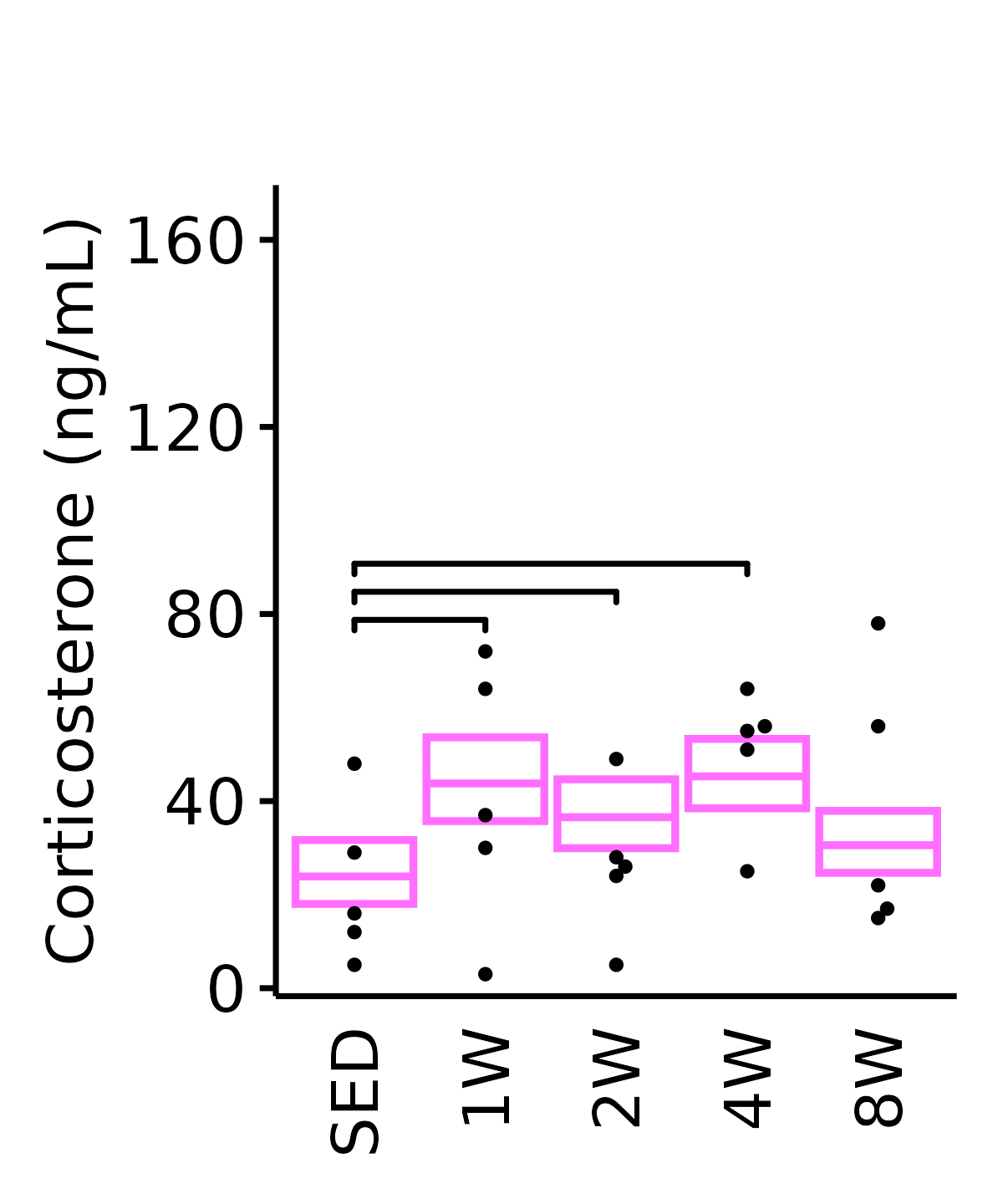
18M Male
plot_baseline(ANALYTES,
response = "corticosterone",
conf = filter(conf_df, response == "Corticosterone"),
stats = filter(ANALYTES_STATS,
response == "Corticosterone"),
sex = "Male", age = "18M") +
scale_y_continuous(name = "Corticosterone (ng/mL)",
breaks = seq(0, 170, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 170), clip = "off") +
theme(plot.margin = unit(c(20, 5, 5, 5), units = "pt"))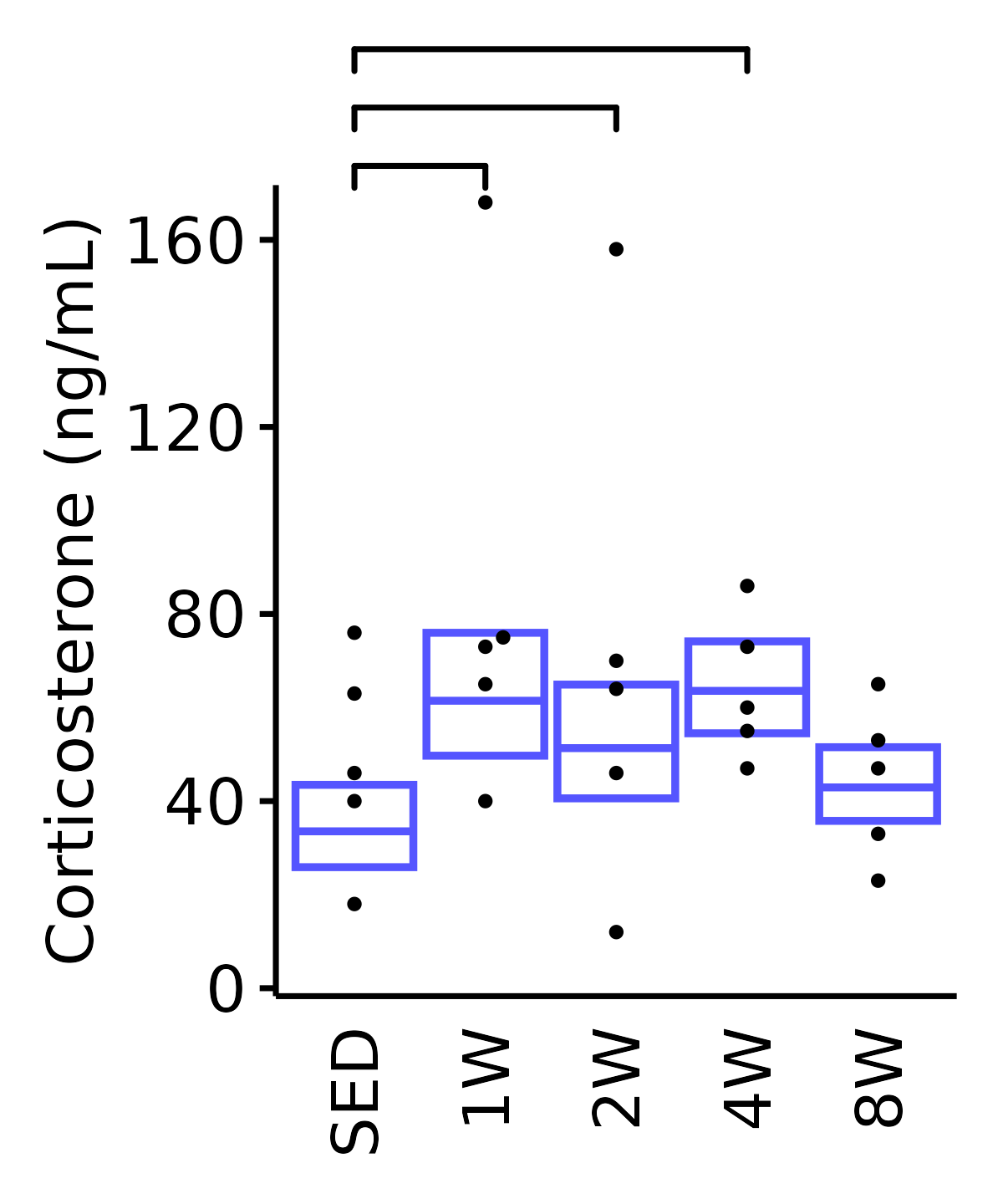
Glucagon
Glucagon (pM).
6M Female
plot_baseline(ANALYTES,
response = "glucagon",
conf = filter(conf_df, response == "Glucagon"),
stats = filter(ANALYTES_STATS, response == "Glucagon"),
sex = "Female", age = "6M") +
scale_y_continuous(name = "Glucagon (pM)",
breaks = seq(0, 160, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 162), clip = "off") +
theme(plot.margin = unit(c(8, 5, 5, 5), units = "pt"))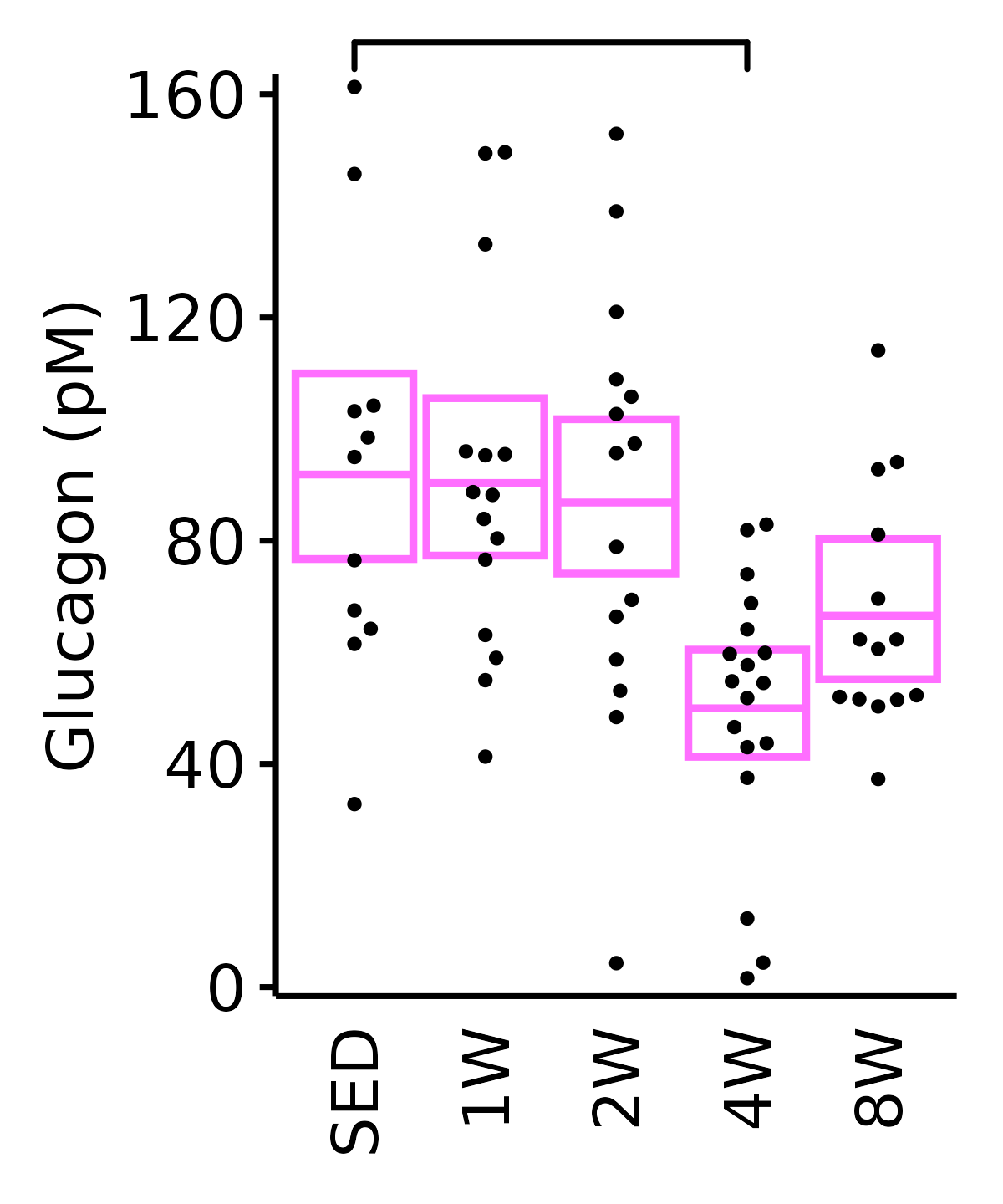
6M Male
plot_baseline(ANALYTES,
response = "glucagon",
conf = filter(conf_df, response == "Glucagon"),
stats = filter(ANALYTES_STATS, response == "Glucagon"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "Glucagon (pM)",
breaks = seq(0, 160, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 162), clip = "off") +
theme(plot.margin = unit(c(8, 5, 5, 5), units = "pt"))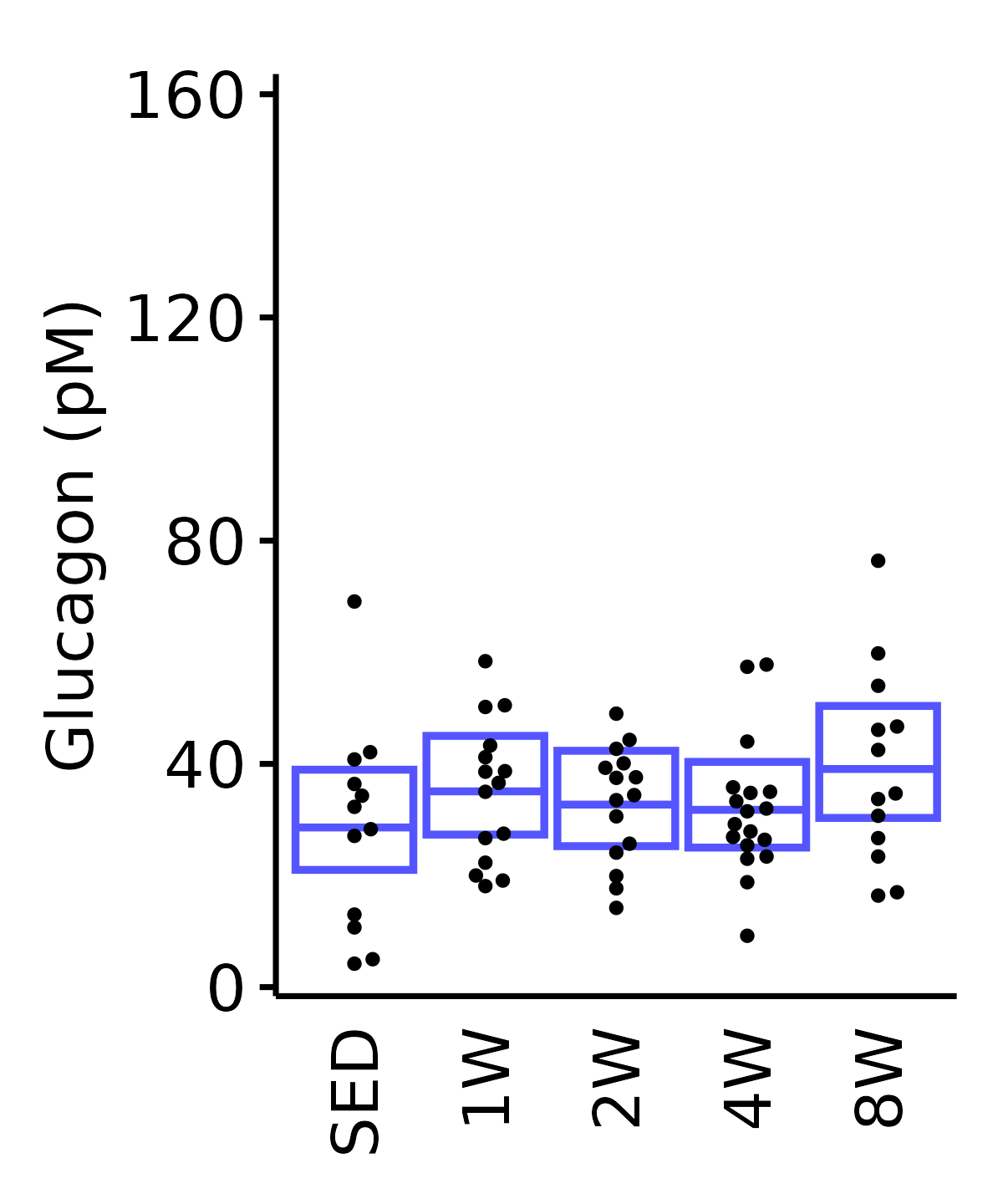
18M Female
plot_baseline(ANALYTES,
response = "glucagon",
conf = filter(conf_df, response == "Glucagon"),
stats = filter(ANALYTES_STATS, response == "Glucagon"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "Glucagon (pM)",
breaks = seq(0, 160, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 162), clip = "off") +
theme(plot.margin = unit(c(8, 5, 5, 5), units = "pt"))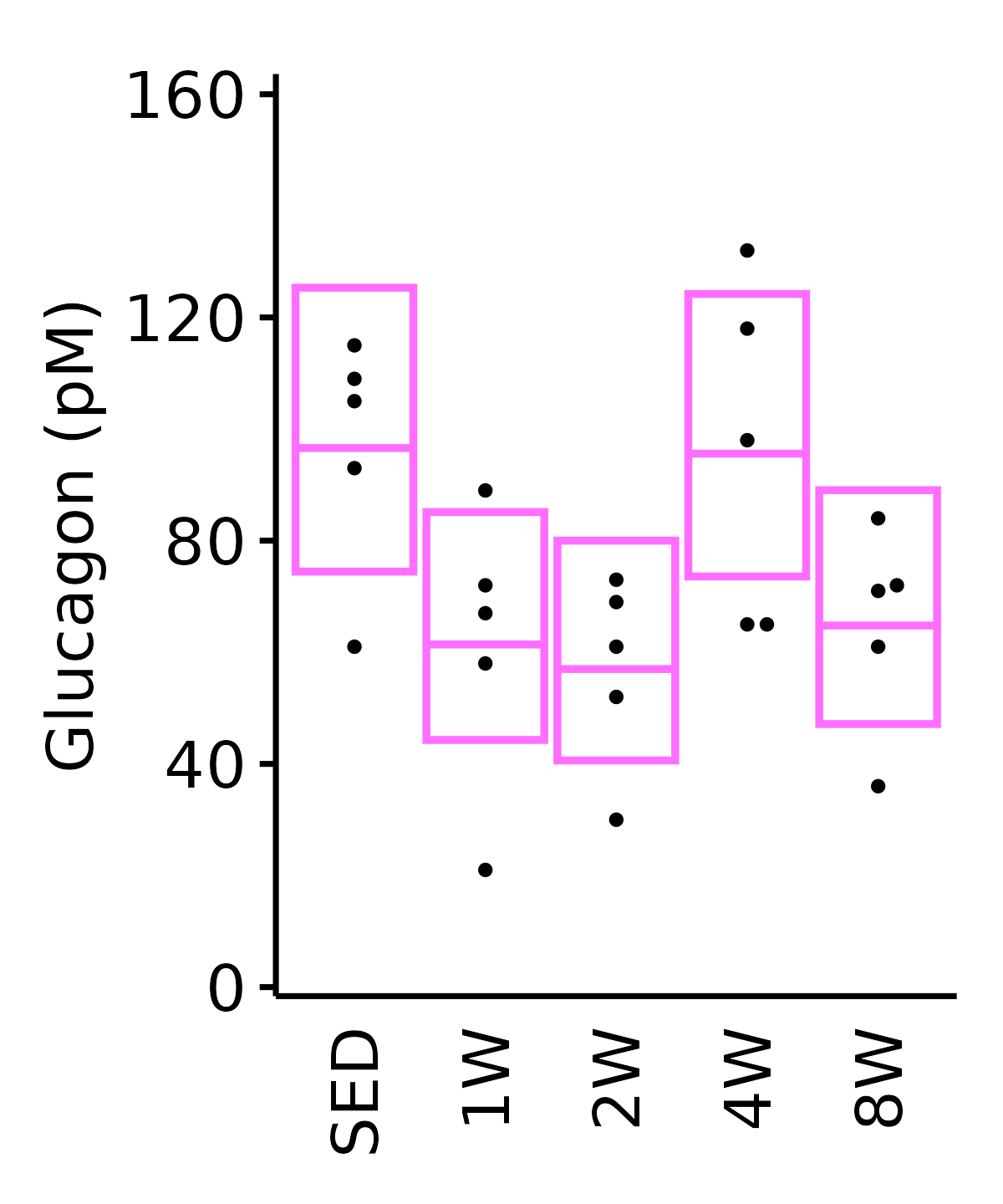
18M Male
plot_baseline(ANALYTES,
response = "glucagon",
conf = filter(conf_df, response == "Glucagon"),
stats = filter(ANALYTES_STATS, response == "Glucagon"),
sex = "Male", age = "18M") +
scale_y_continuous(name = "Glucagon (pM)",
breaks = seq(0, 160, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 162), clip = "off") +
theme(plot.margin = unit(c(8, 5, 5, 5), units = "pt"))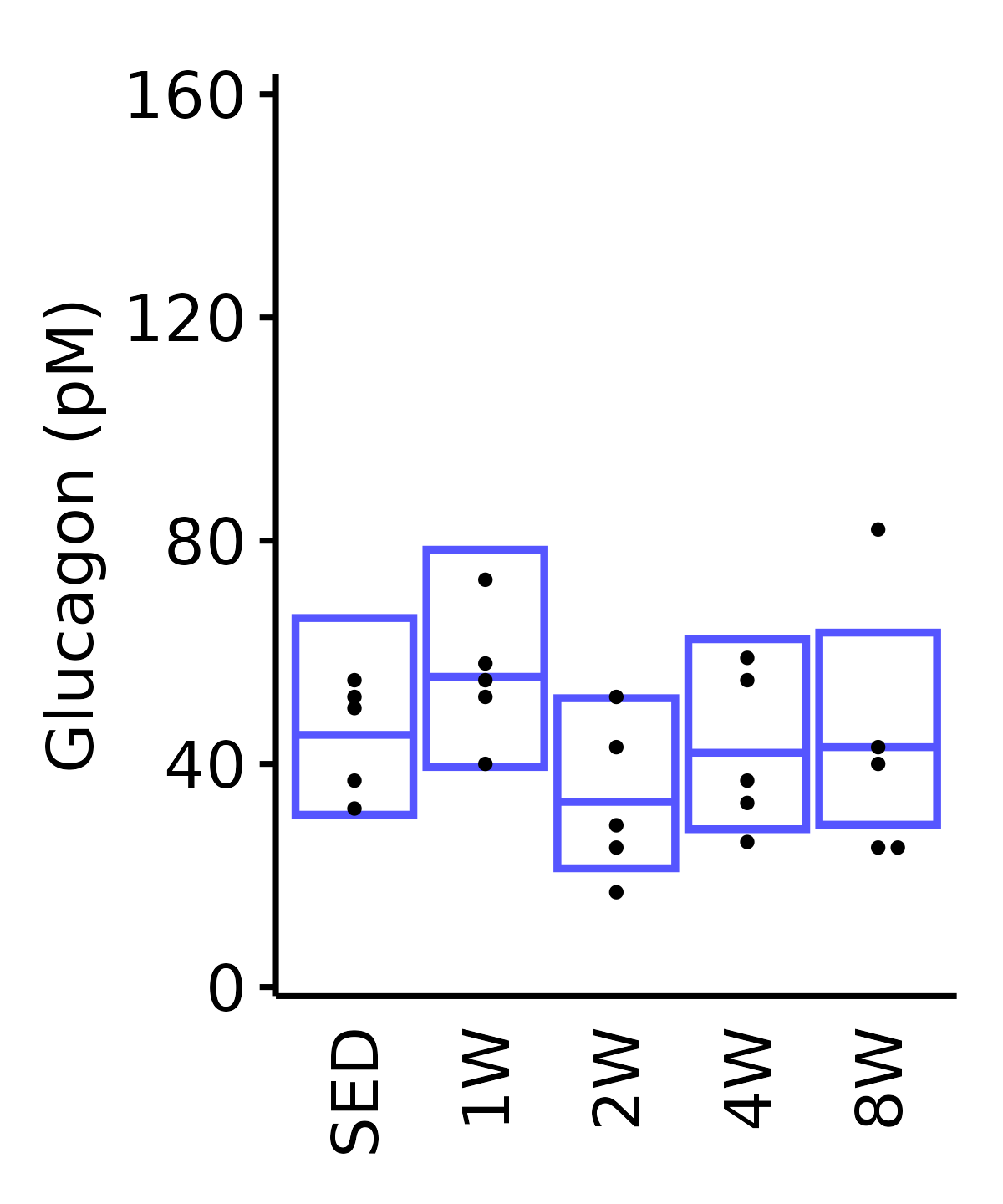
Glucose
Glucose (mg/dL).
6M Female
plot_baseline(ANALYTES,
response = "glucose",
conf = filter(conf_df, response == "Glucose"),
stats = filter(ANALYTES_STATS, response == "Glucose"),
sex = "Female", age = "6M") +
scale_y_continuous(name = "Glucose (mg/dL)",
breaks = seq(110, 230, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(110, 230), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))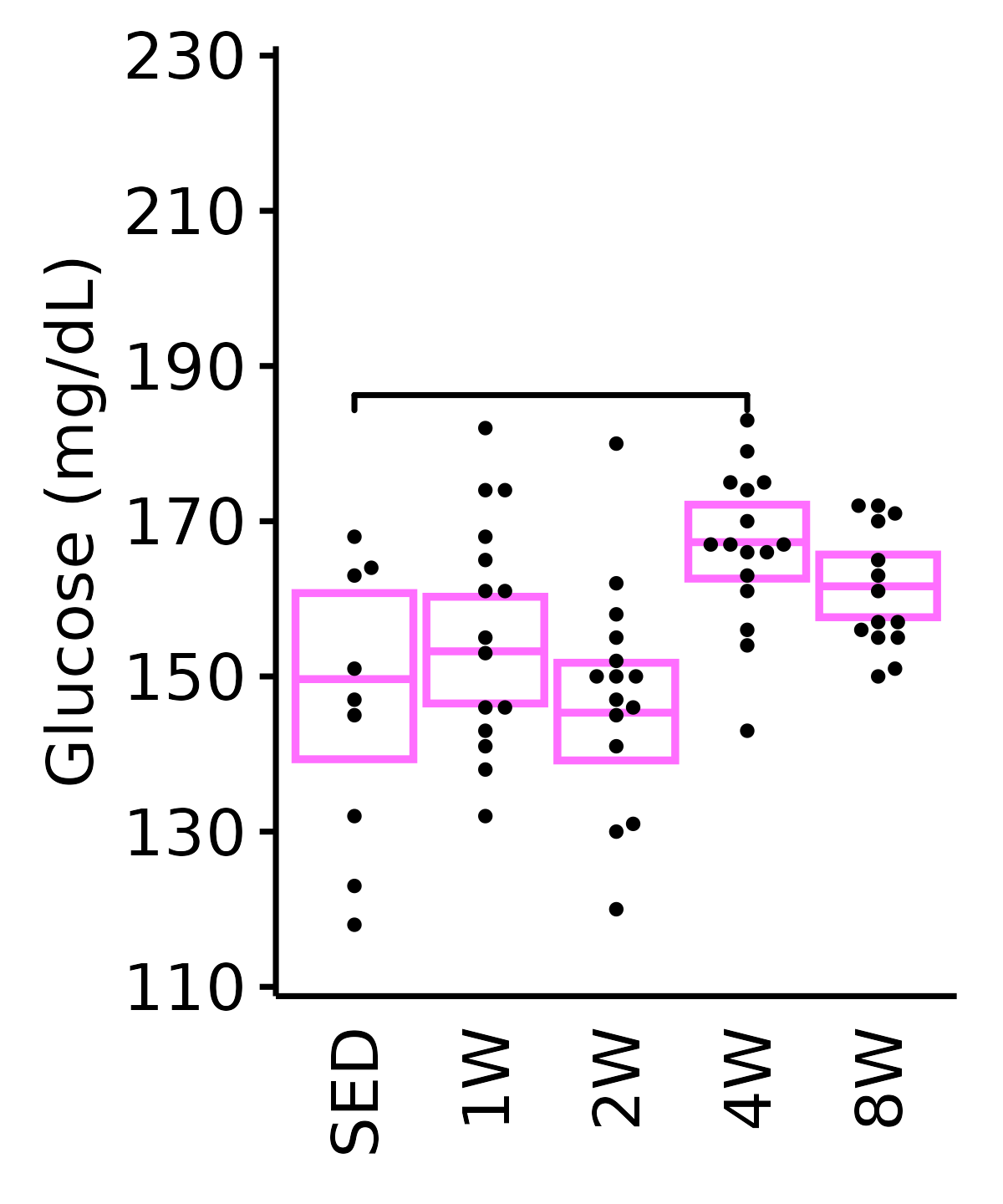
6M Male
plot_baseline(ANALYTES,
response = "glucose",
conf = filter(conf_df, response == "Glucose"),
stats = filter(ANALYTES_STATS, response == "Glucose"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "Glucose (mg/dL)",
breaks = seq(110, 230, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(110, 230), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))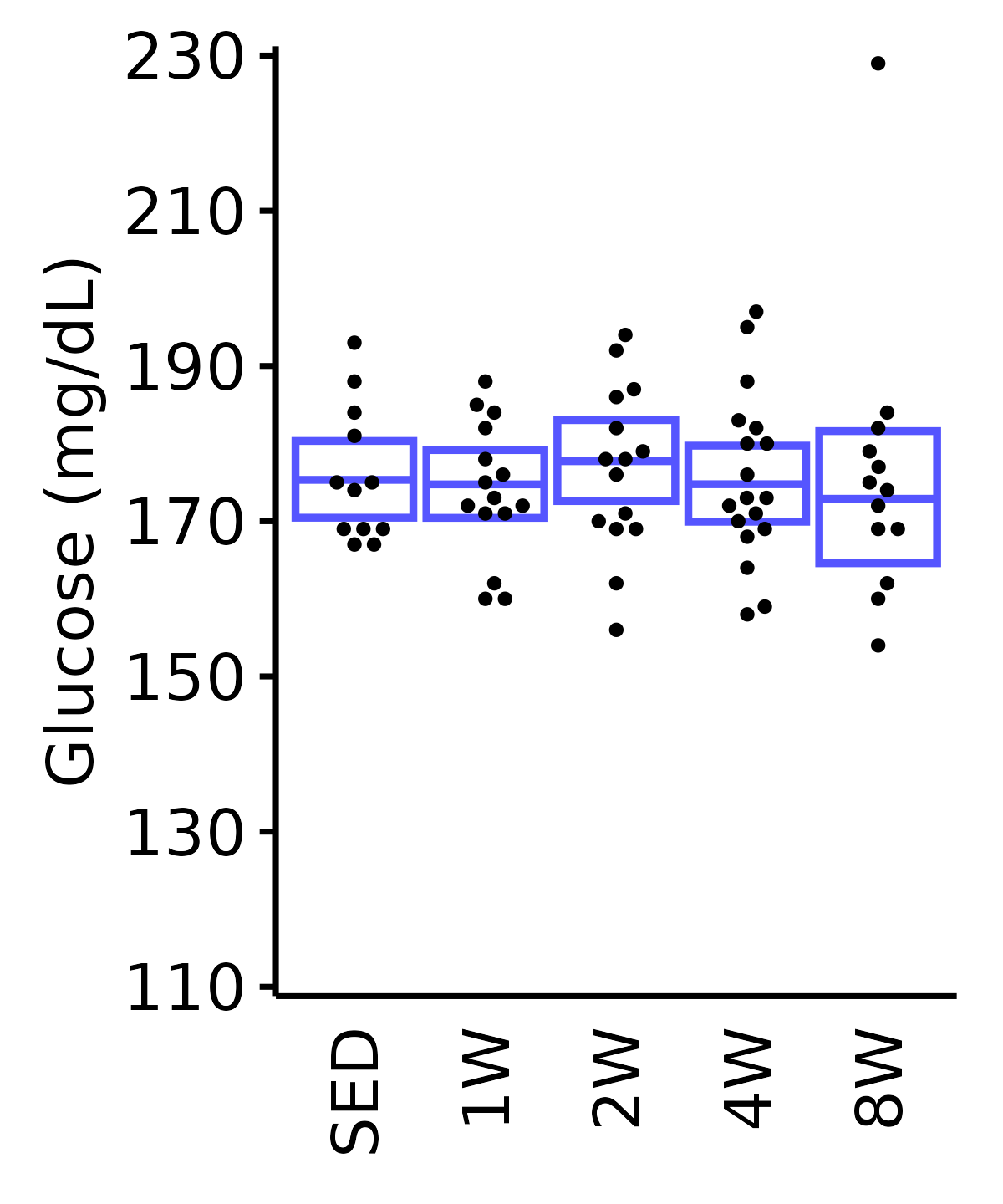
18M Female
plot_baseline(ANALYTES,
response = "glucose",
conf = filter(conf_df, response == "Glucose"),
stats = filter(ANALYTES_STATS, response == "Glucose"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "Glucose (mg/dL)",
breaks = seq(110, 230, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(110, 230), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))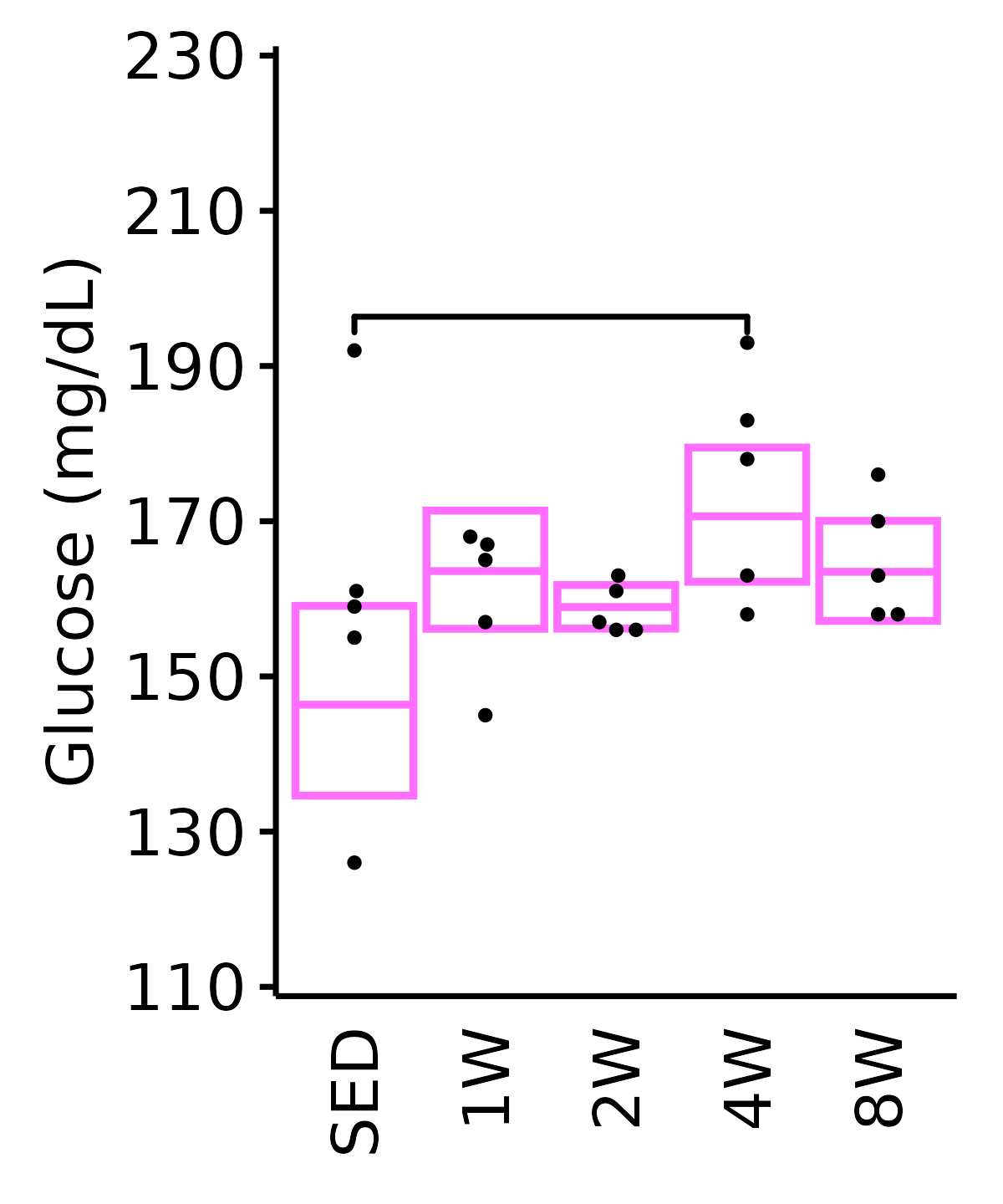
18M Male
plot_baseline(ANALYTES,
response = "glucose",
conf = filter(conf_df, response == "Glucose"),
stats = filter(ANALYTES_STATS, response == "Glucose"),
sex = "Male", age = "18M", y_position = 204) +
scale_y_continuous(name = "Glucose (mg/dL)",
breaks = seq(110, 230, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(110, 230), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))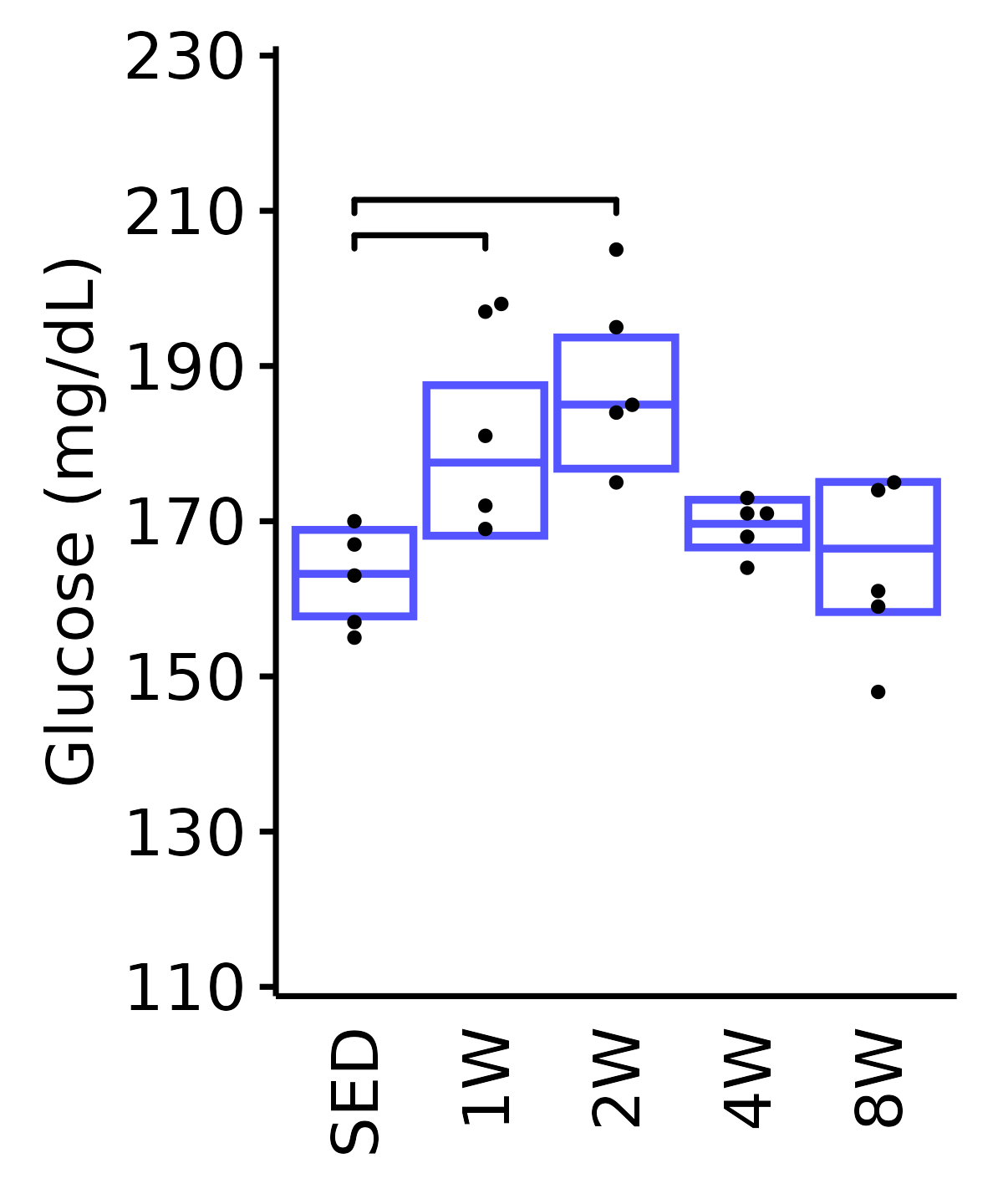
Glycerol
6M Female
plot_baseline(ANALYTES,
response = "glycerol",
conf = filter(conf_df, response == "Glycerol"),
stats = filter(ANALYTES_STATS, response == "Glycerol"),
sex = "Female", age = "6M") +
scale_y_continuous(name = "Glycerol (mg/dL)",
breaks = seq(0, 2.5, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 2.7), clip = "off") +
theme(plot.margin = unit(c(10, 5, 5, 5), units = "pt"))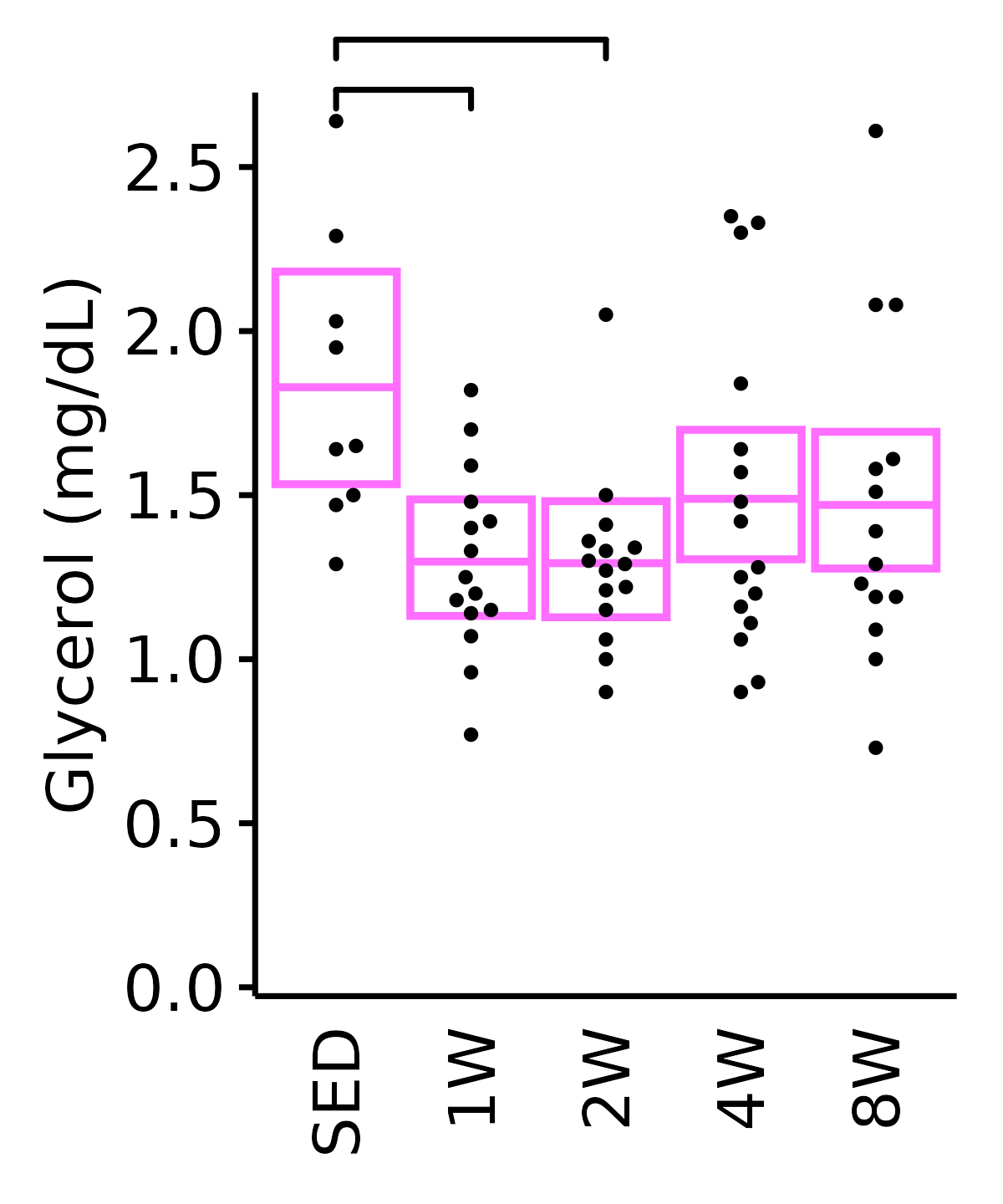
6M Male
plot_baseline(ANALYTES,
response = "glycerol",
conf = filter(conf_df, response == "Glycerol"),
stats = filter(ANALYTES_STATS, response == "Glycerol"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "Glycerol (mg/dL)",
breaks = seq(0, 2.5, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 2.7), clip = "off") +
theme(plot.margin = unit(c(10, 5, 5, 5), units = "pt"))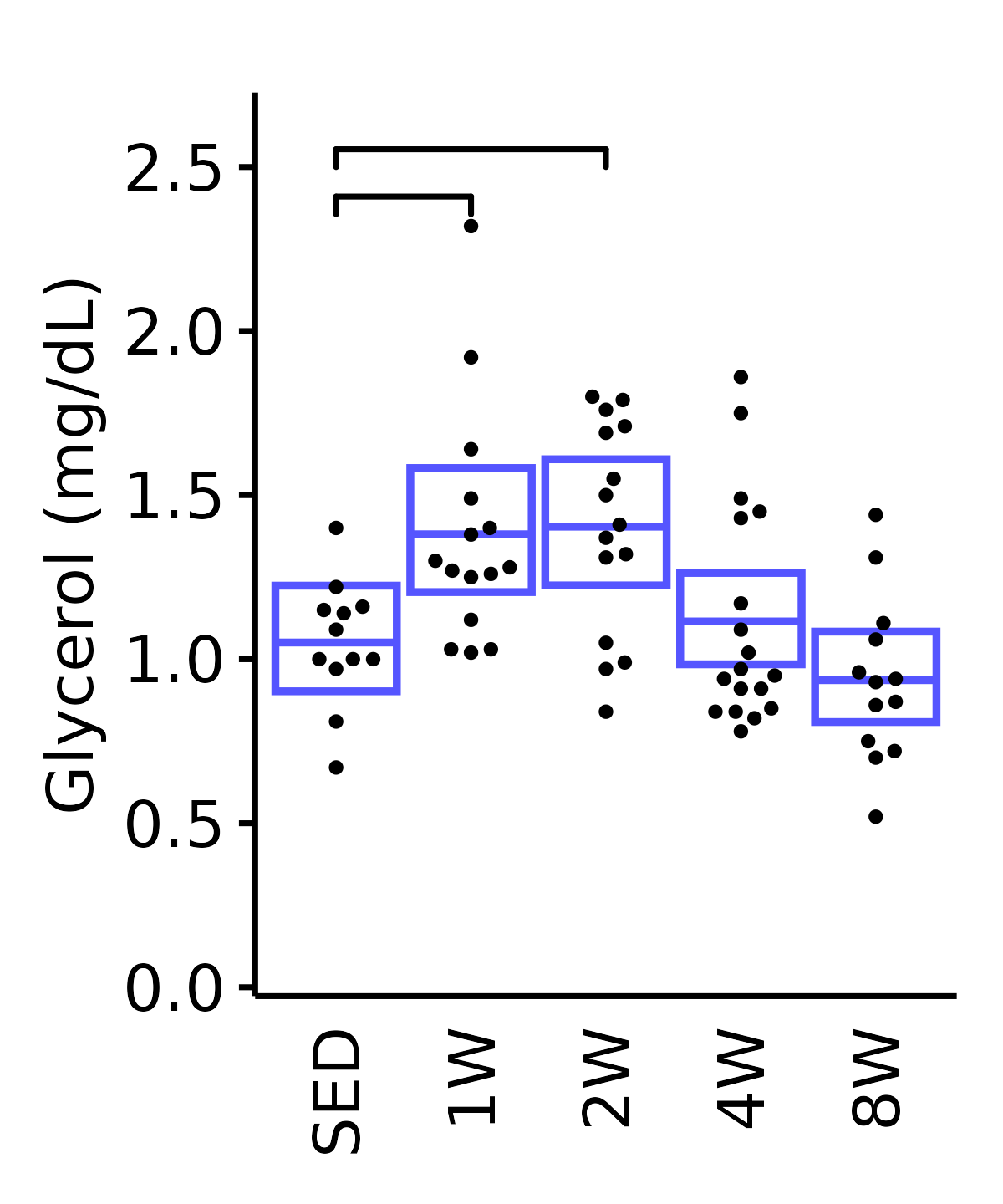
18M Female
plot_baseline(ANALYTES,
response = "glycerol",
conf = filter(conf_df, response == "Glycerol"),
stats = filter(ANALYTES_STATS, response == "Glycerol"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "Glycerol (mg/dL)",
breaks = seq(0, 2.5, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 2.7), clip = "off") +
theme(plot.margin = unit(c(10, 5, 5, 5), units = "pt"))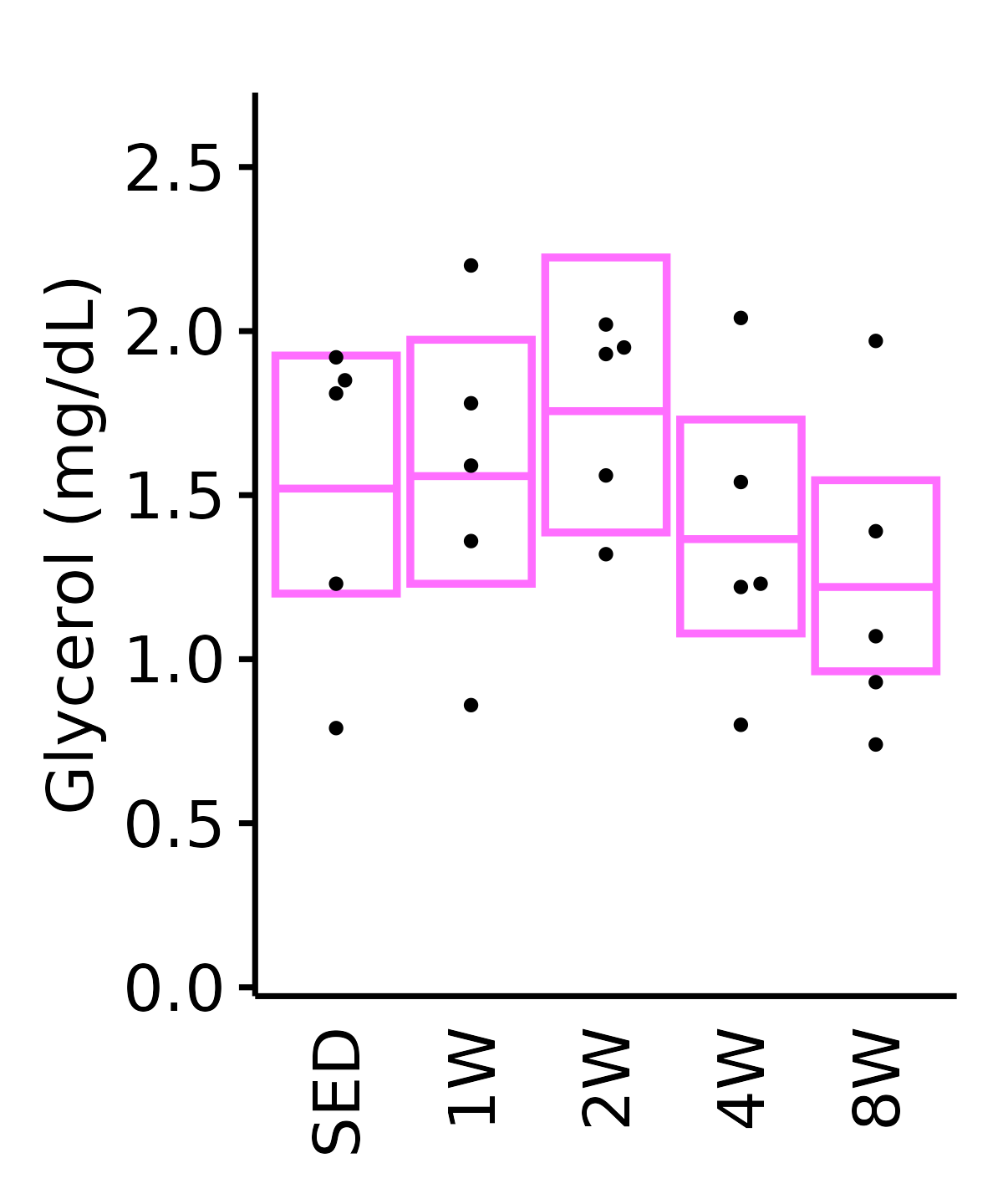
18M Male
plot_baseline(ANALYTES,
response = "glycerol",
conf = filter(conf_df, response == "Glycerol"),
stats = filter(ANALYTES_STATS, response == "Glycerol"),
sex = "Male", age = "18M") +
scale_y_continuous(name = "Glycerol (mg/dL)",
breaks = seq(0, 2.5, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 2.7), clip = "off") +
theme(plot.margin = unit(c(10, 5, 5, 5), units = "pt"))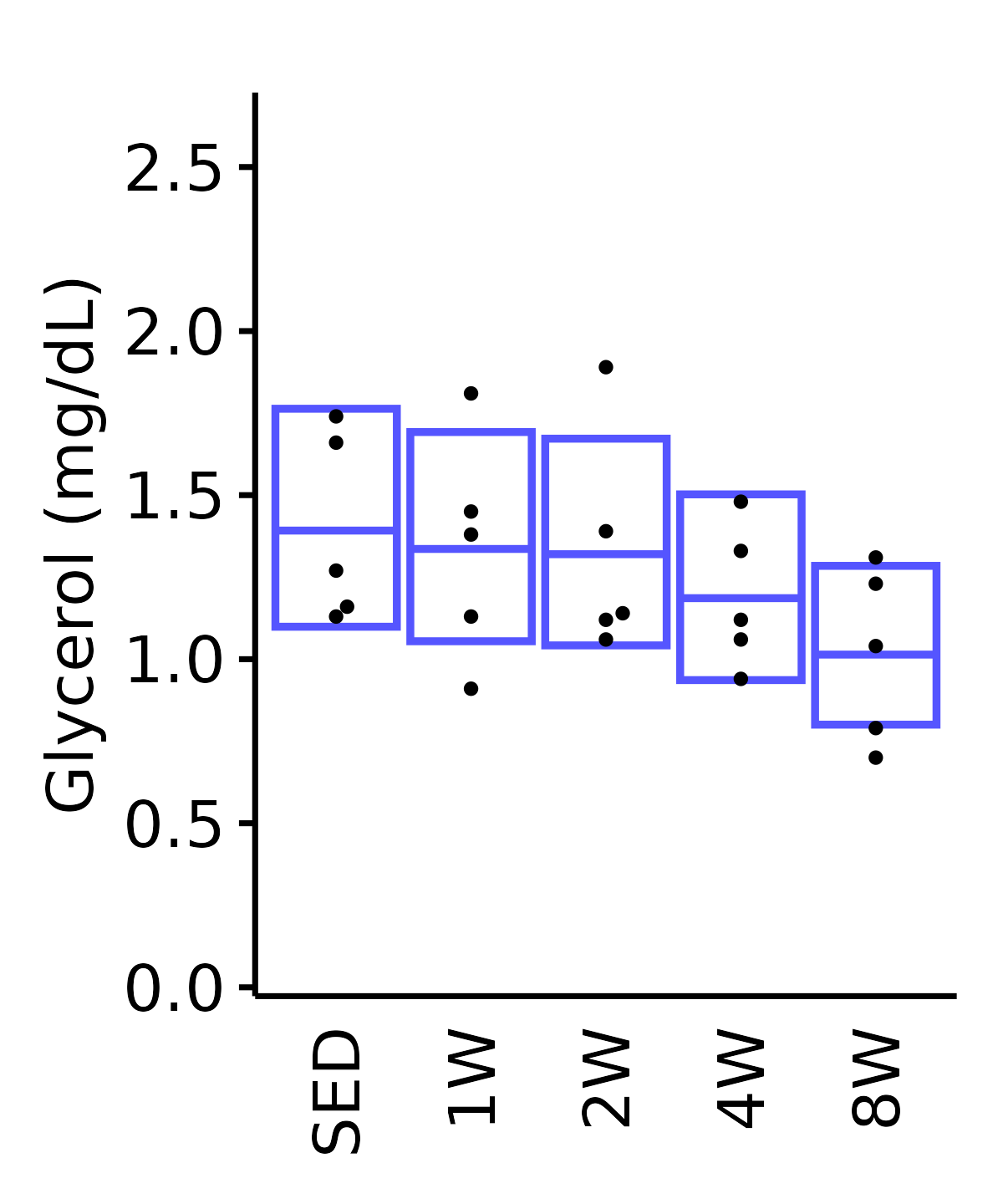
Insulin
6M Female
plot_baseline(ANALYTES,
response = "insulin_pm",
conf = filter(conf_df, response == "Insulin"),
stats = filter(ANALYTES_STATS, response == "Insulin"),
sex = "Female", age = "6M") +
scale_y_continuous(name = "Insulin (pM)",
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1400), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))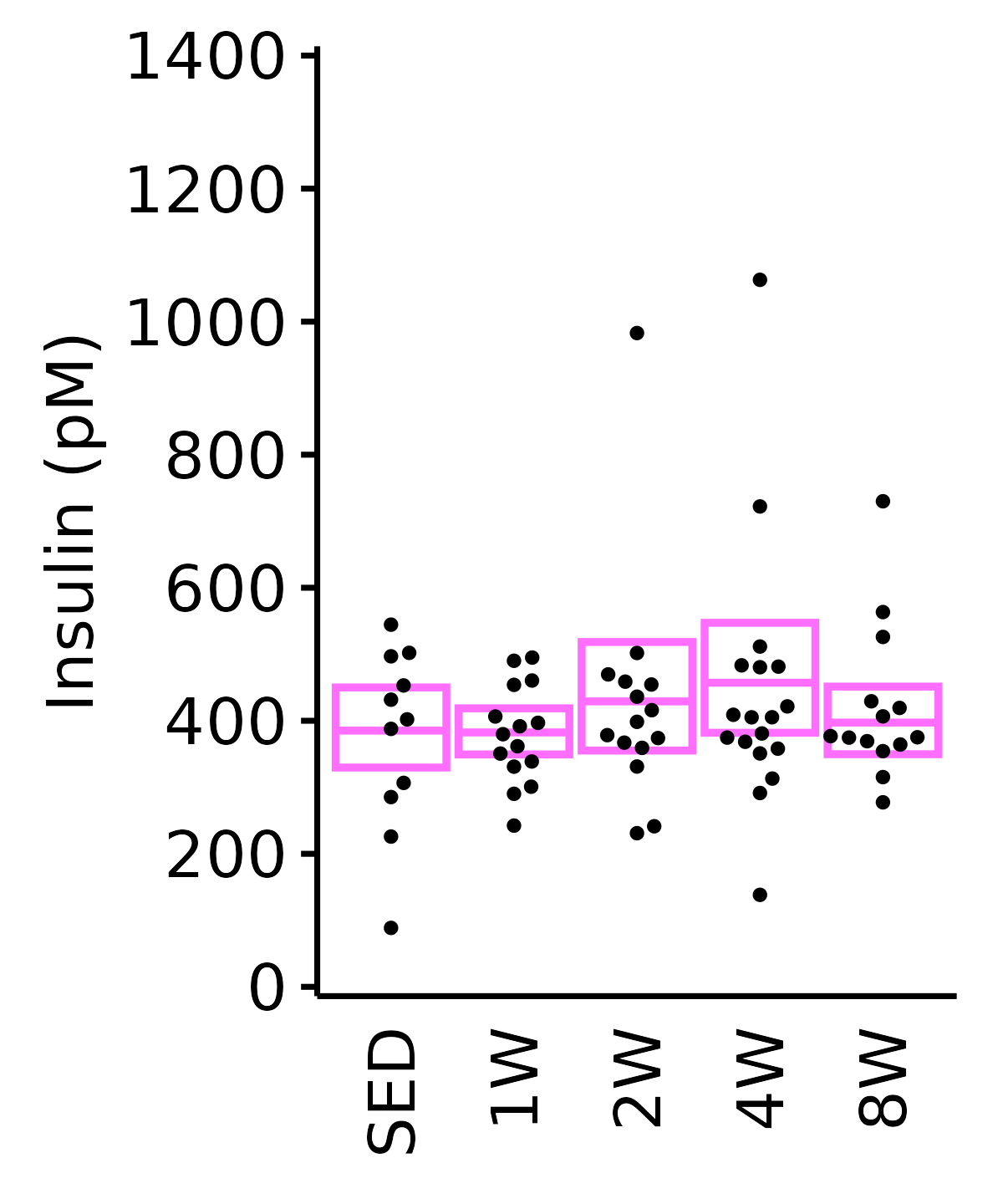
6M Male
plot_baseline(ANALYTES,
response = "insulin_pm",
conf = filter(conf_df, response == "Insulin"),
stats = filter(ANALYTES_STATS, response == "Insulin"),
sex = "Male", age = "6M", y_position = 970) +
scale_y_continuous(name = "Insulin (pM)",
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1400), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))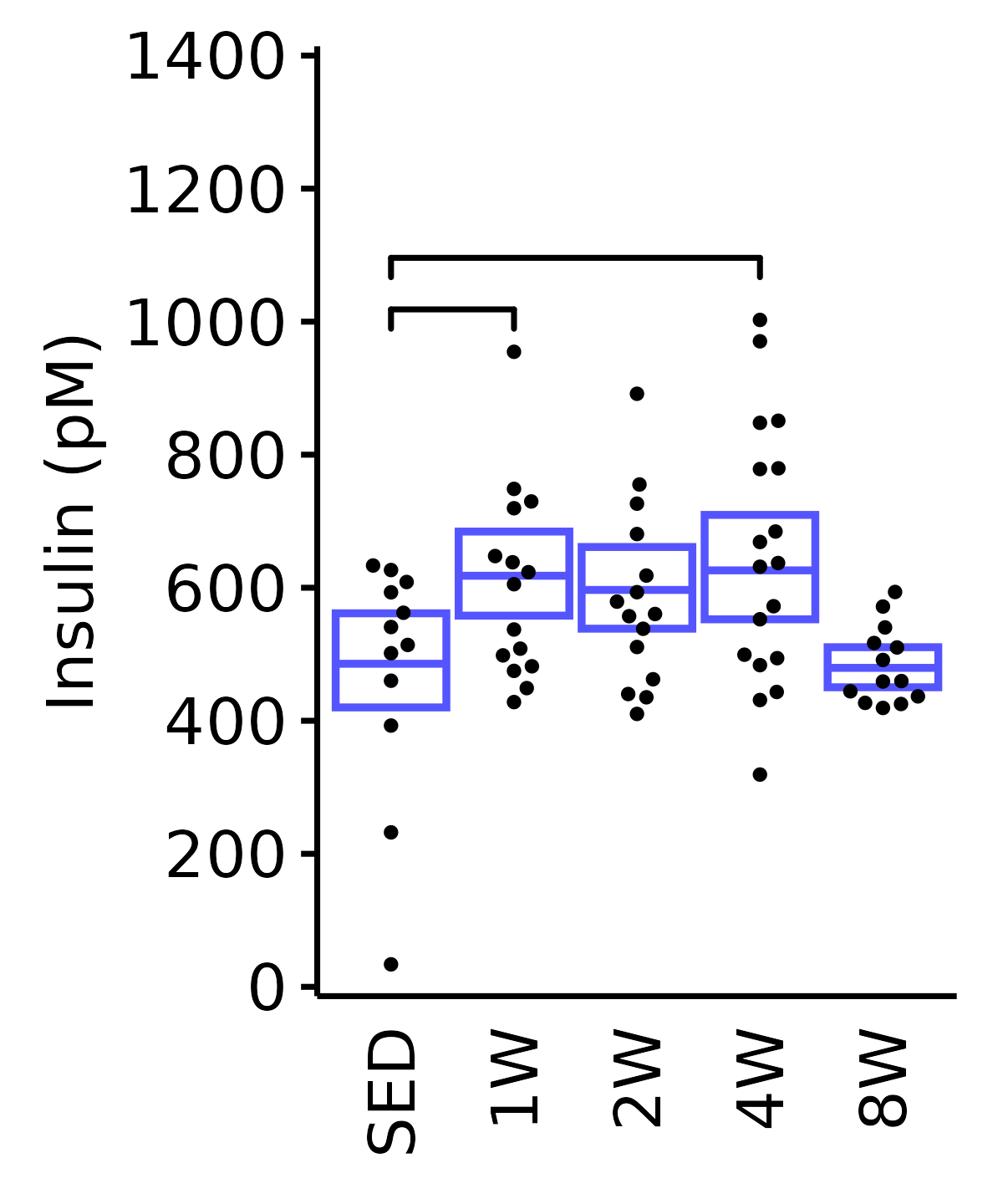
18M Female
plot_baseline(ANALYTES,
response = "insulin_pm",
conf = filter(conf_df, response == "Insulin"),
stats = filter(ANALYTES_STATS, response == "Insulin"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "Insulin (pM)",
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1400), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))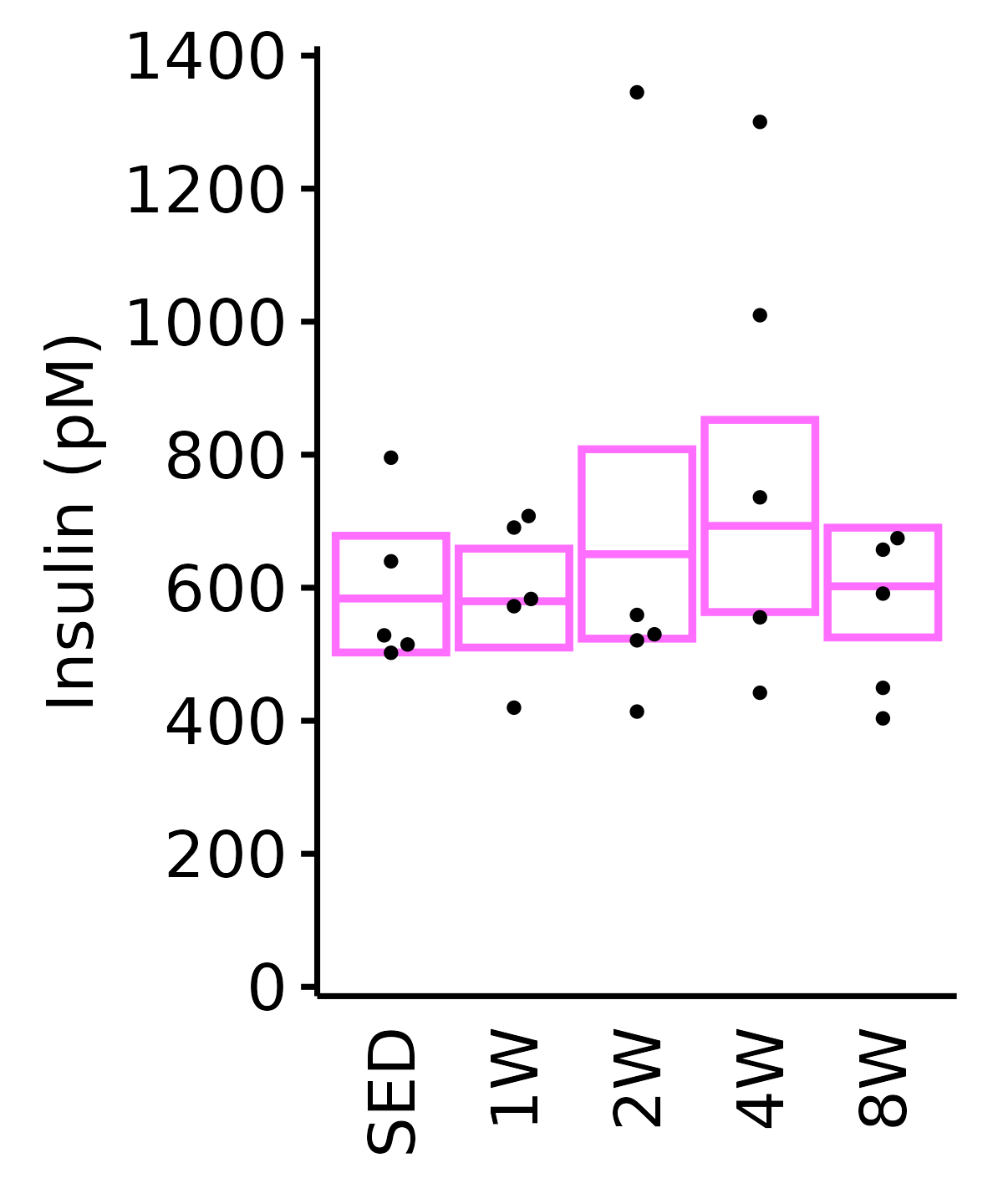
18M Male
plot_baseline(ANALYTES,
response = "insulin_pm",
conf = filter(conf_df, response == "Insulin"),
stats = filter(ANALYTES_STATS, response == "Insulin"),
sex = "Male", age = "18M", y_position = 940) +
scale_y_continuous(name = "Insulin (pM)",
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1400), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))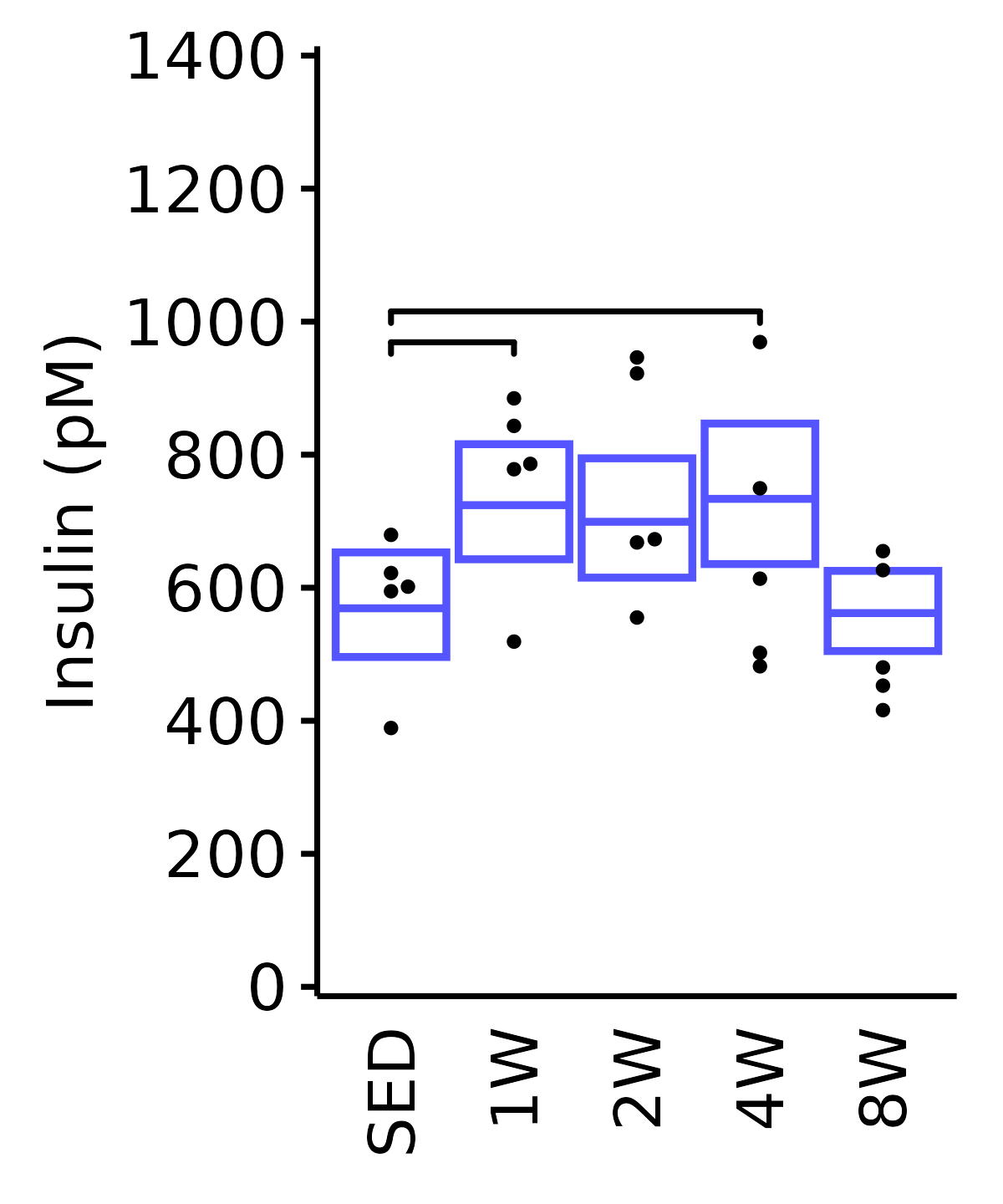
Ketones
Ketones (\(\mu\)mol/L).
6M Female
plot_baseline(ANALYTES,
response = "ketones",
conf = filter(conf_df, response == "Ketones"),
stats = filter(ANALYTES_STATS, response == "Ketones"),
sex = "Female", age = "6M") +
scale_y_continuous(name = TeX("Ketones ($\\mu$mol/L)"),
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1450), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))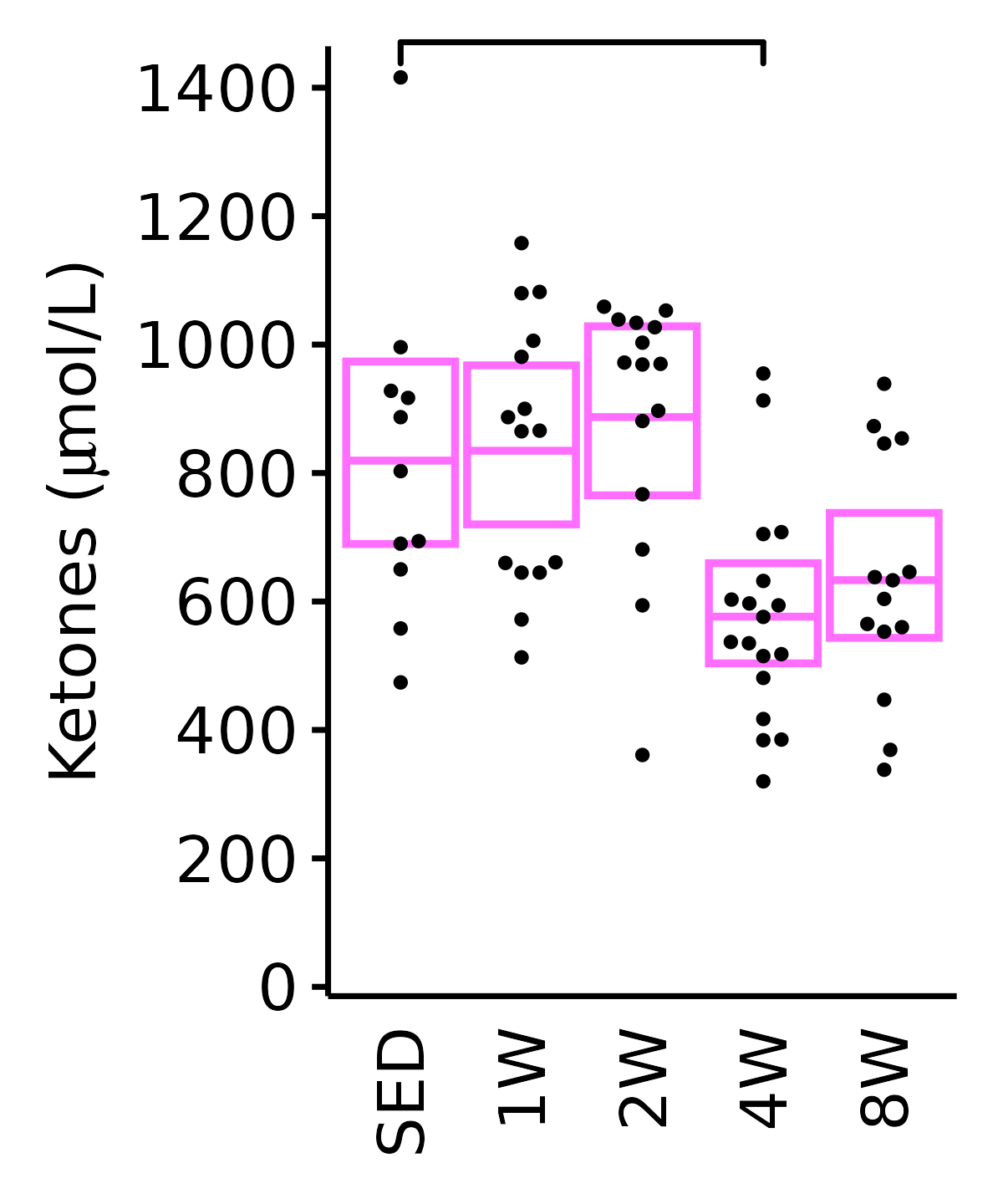
6M Male
plot_baseline(ANALYTES,
response = "ketones",
conf = filter(conf_df, response == "Ketones"),
stats = filter(ANALYTES_STATS, response == "Ketones"),
sex = "Male", age = "6M") +
scale_y_continuous(name = TeX("Ketones ($\\mu$mol/L)"),
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1450), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))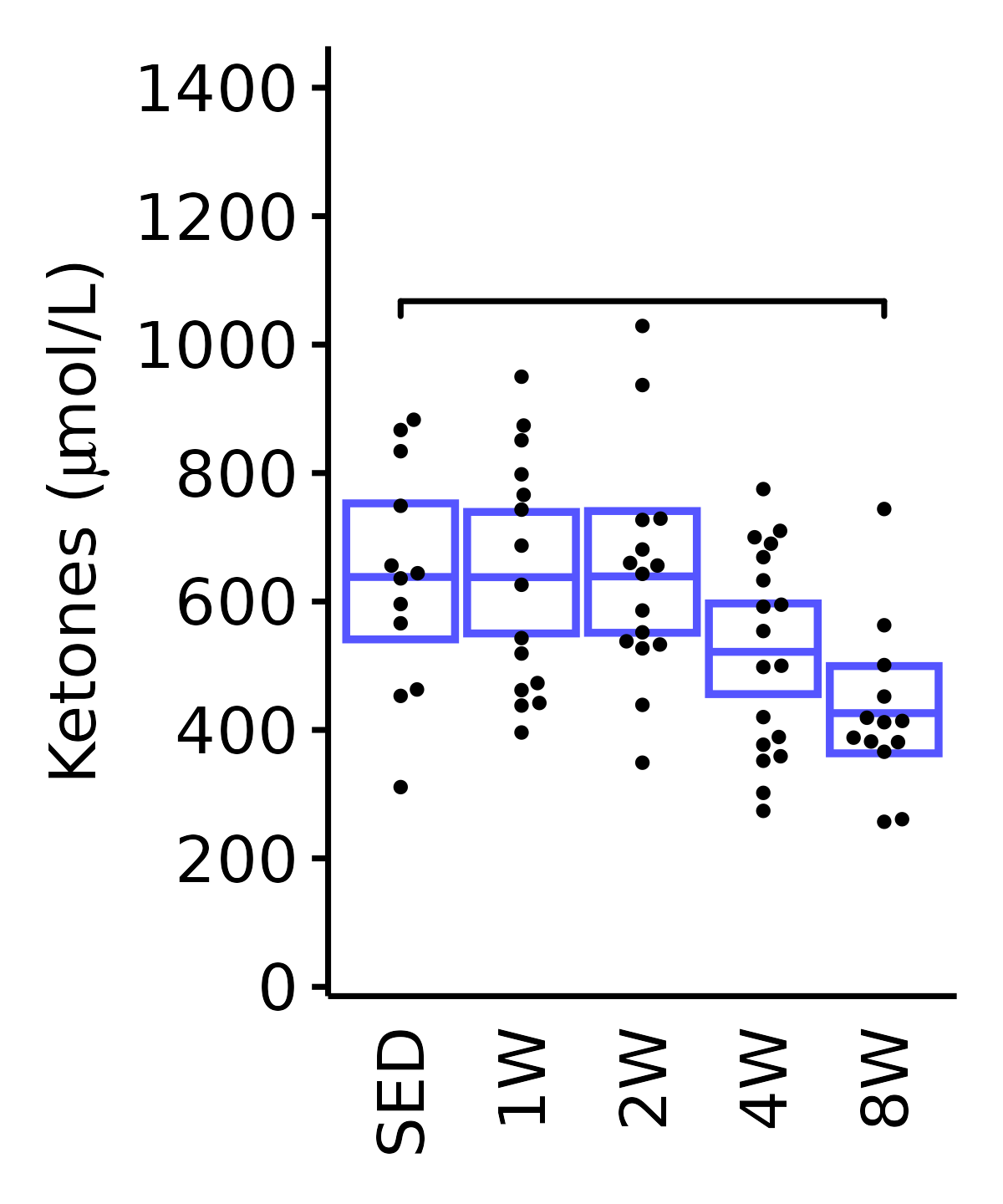
18M Female
plot_baseline(ANALYTES,
response = "ketones",
conf = filter(conf_df, response == "Ketones"),
stats = filter(ANALYTES_STATS, response == "Ketones"),
sex = "Female", age = "18M") +
labs(y = TeX("Ketones ($\\mu$mol/L)")) +
scale_y_continuous(name = TeX("Ketones ($\\mu$mol/L)"),
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1450), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))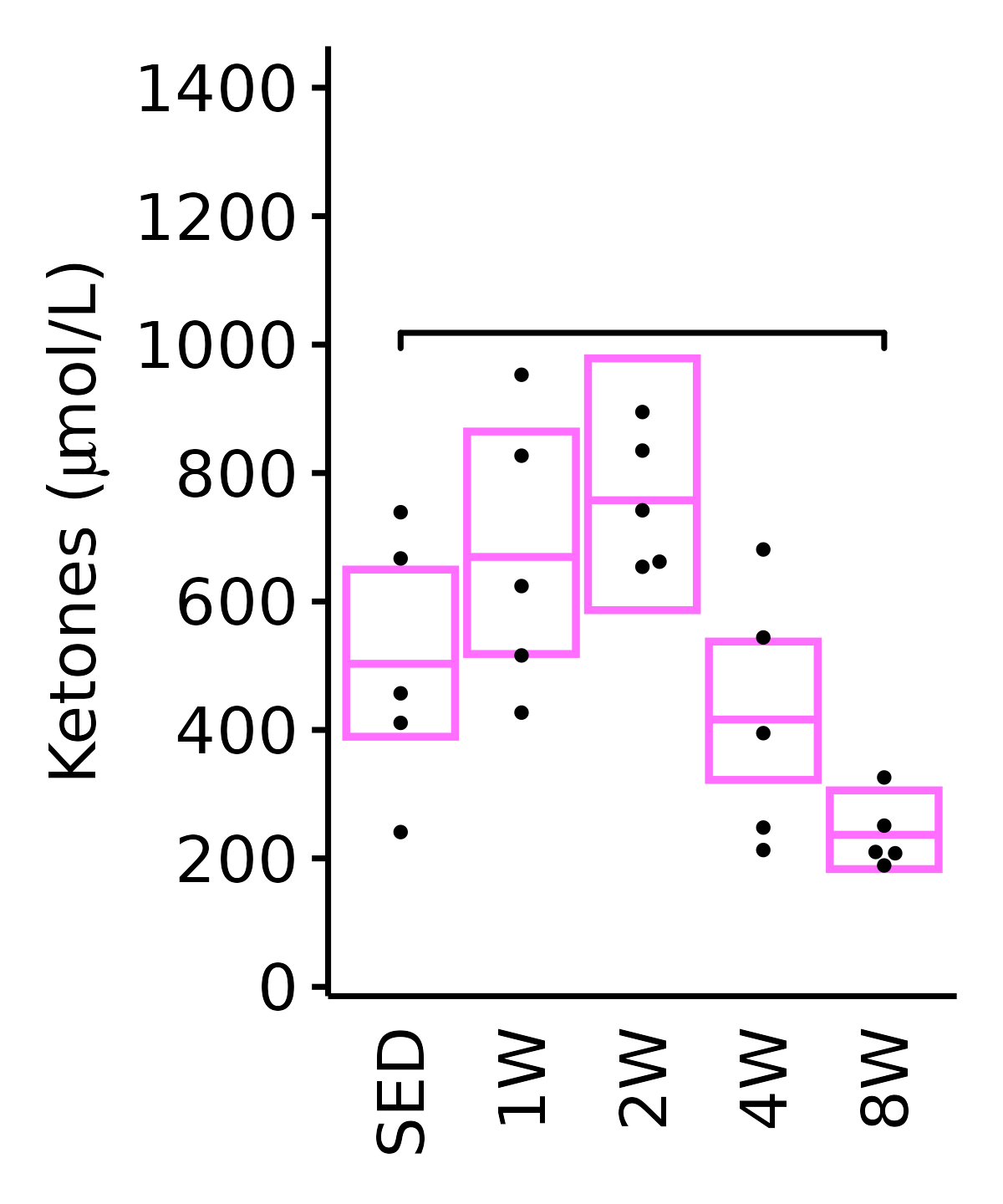
18M Male
plot_baseline(ANALYTES,
response = "ketones",
conf = filter(conf_df, response == "Ketones"),
stats = filter(ANALYTES_STATS, response == "Ketones"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("Ketones ($\\mu$mol/L)"),
breaks = seq(0, 1400, 200),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1450), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))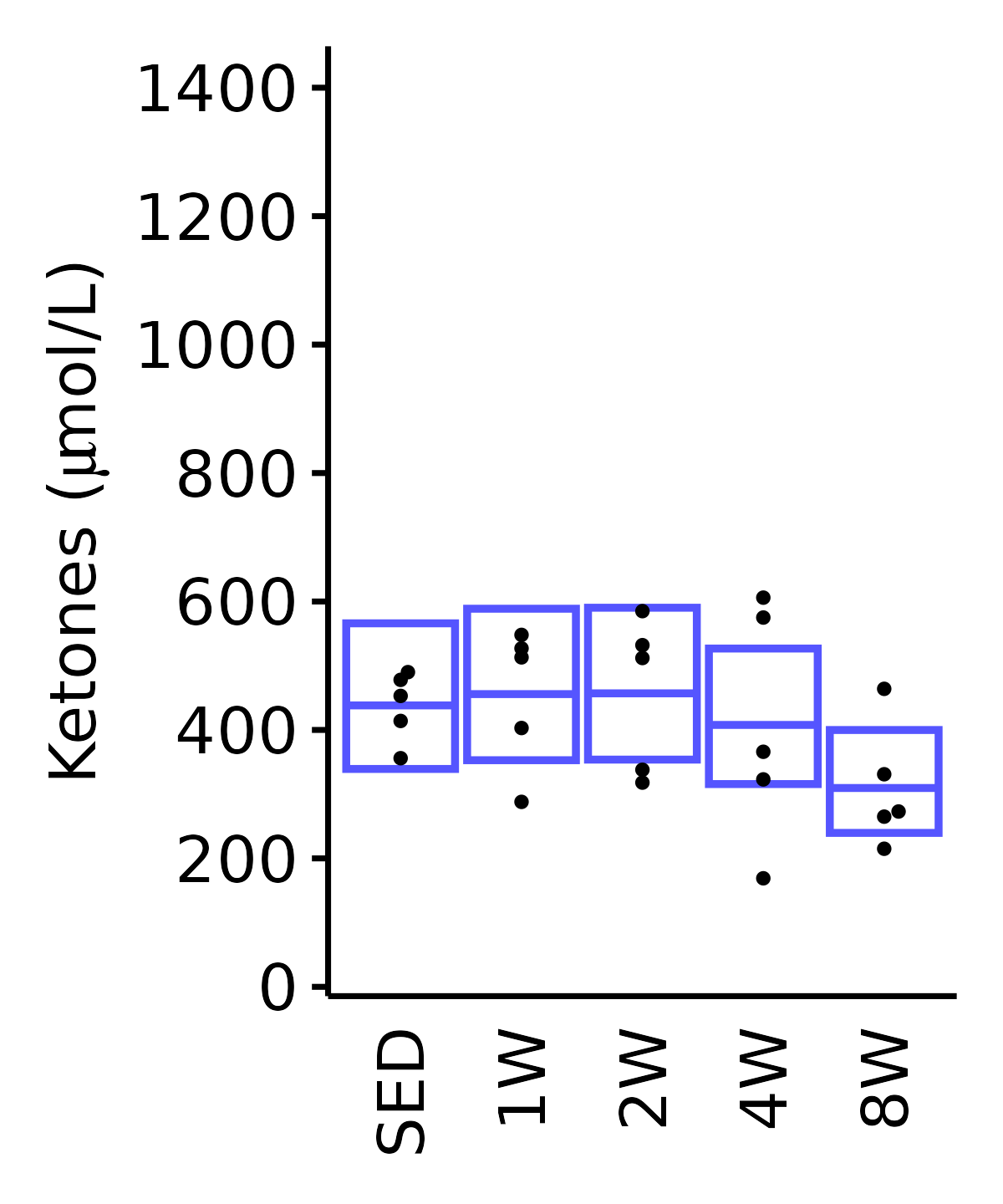
Lactate
Lactate (mmol/L).
6M Female
plot_baseline(ANALYTES,
response = "lactate",
conf = filter(conf_df, response == "Lactate"),
stats = filter(ANALYTES_STATS, response == "Lactate"),
sex = "Female", age = "6M", y_position = 1.85) +
scale_y_continuous(name = "Lactate (mmol/L)",
breaks = seq(0, 3, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 3), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))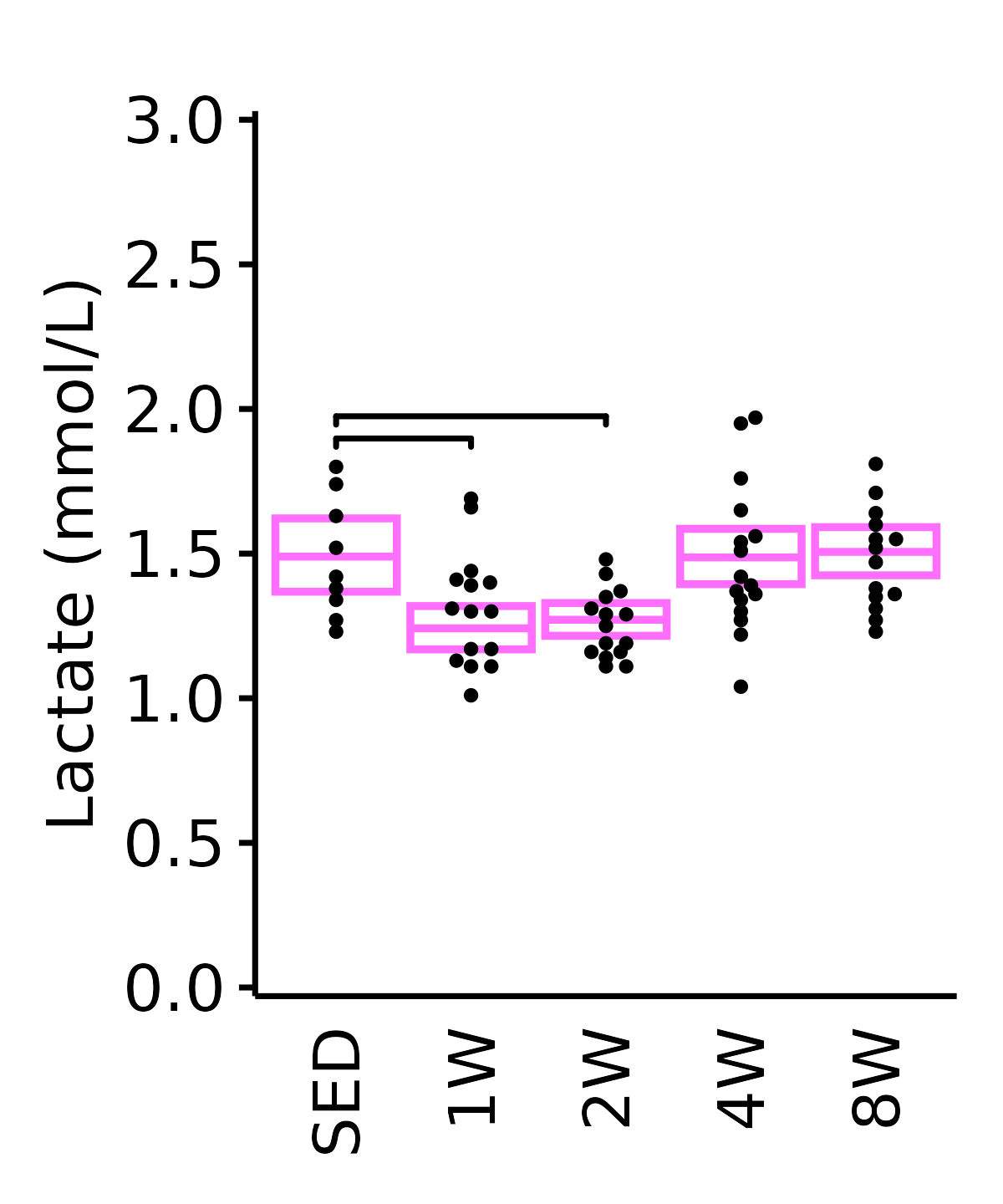
6M Male
plot_baseline(ANALYTES,
response = "lactate",
conf = filter(conf_df, response == "Lactate"),
stats = filter(ANALYTES_STATS, response == "Lactate"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "Lactate (mmol/L)",
breaks = seq(0, 3, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 3), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))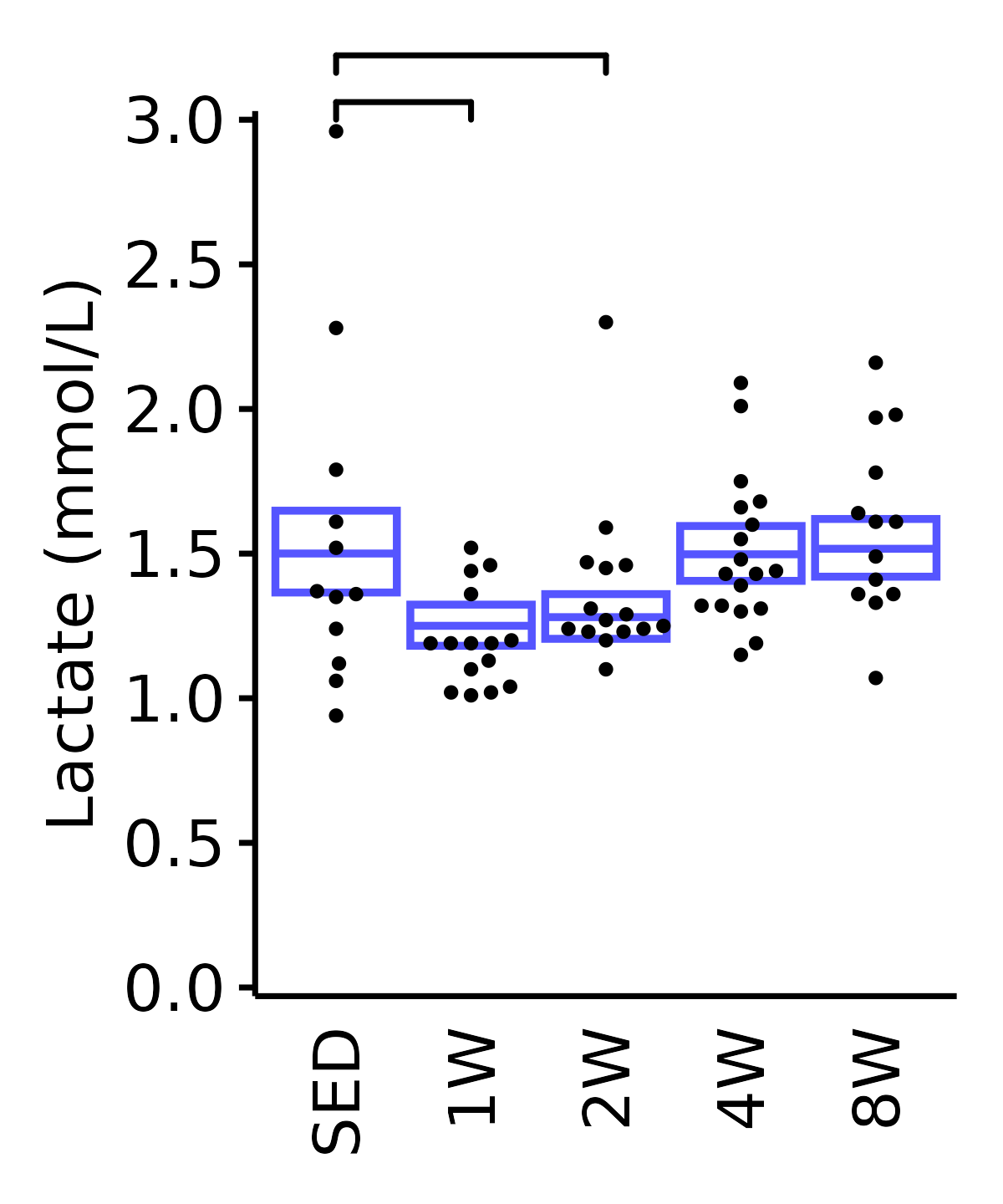
18M Female
plot_baseline(ANALYTES,
response = "lactate",
conf = filter(conf_df, response == "Lactate"),
stats = filter(ANALYTES_STATS, response == "Lactate"),
sex = "Female", age = "18M", y_position = 2.25) +
scale_y_continuous(name = "Lactate (mmol/L)",
breaks = seq(0, 3, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 3), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))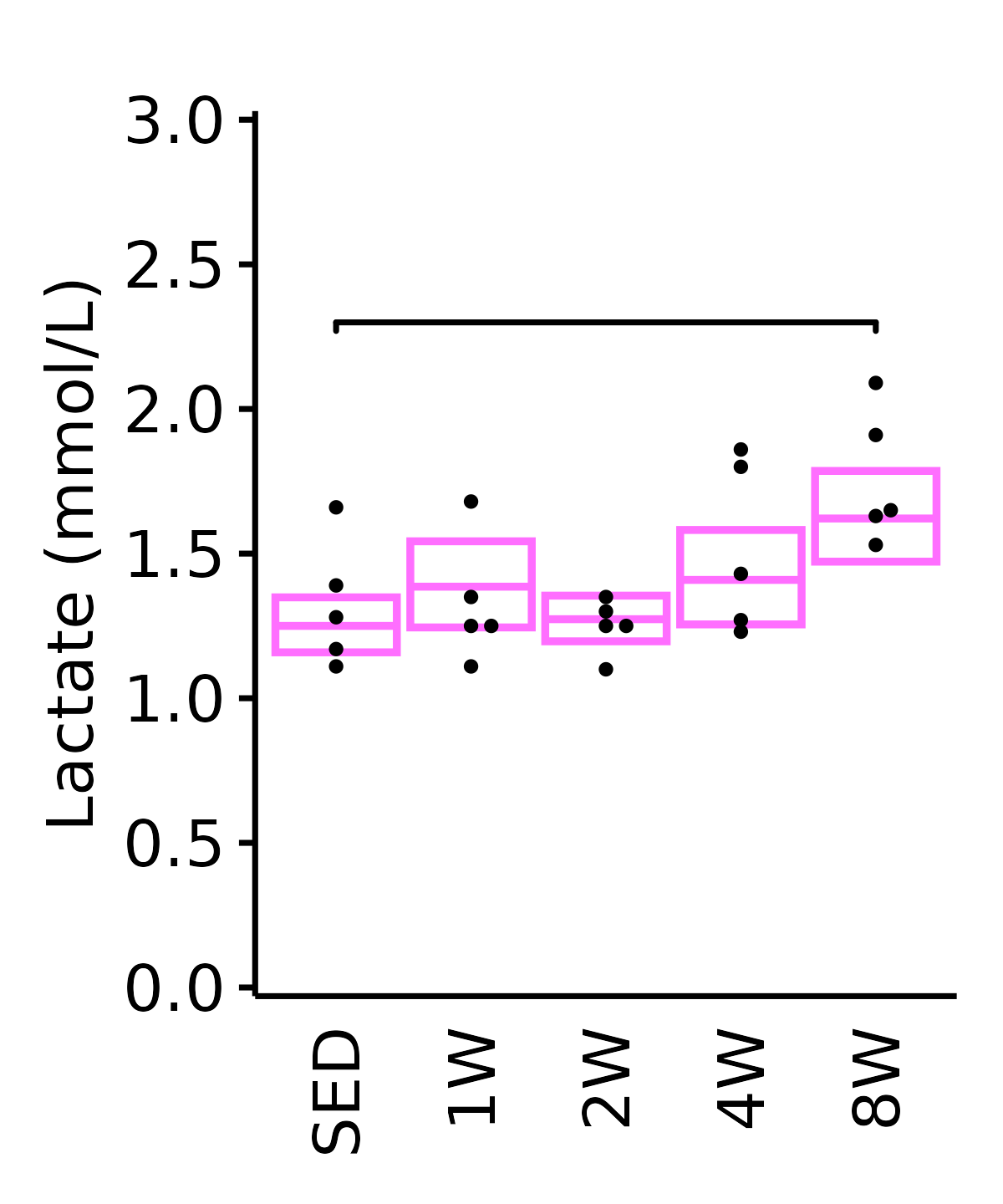
18M Male
plot_baseline(ANALYTES,
response = "lactate",
conf = filter(conf_df, response == "Lactate"),
stats = filter(ANALYTES_STATS, response == "Lactate"),
sex = "Male", age = "18M", y_position = 2) +
scale_y_continuous(name = "Lactate (mmol/L)",
breaks = seq(0, 3, 0.5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 3), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))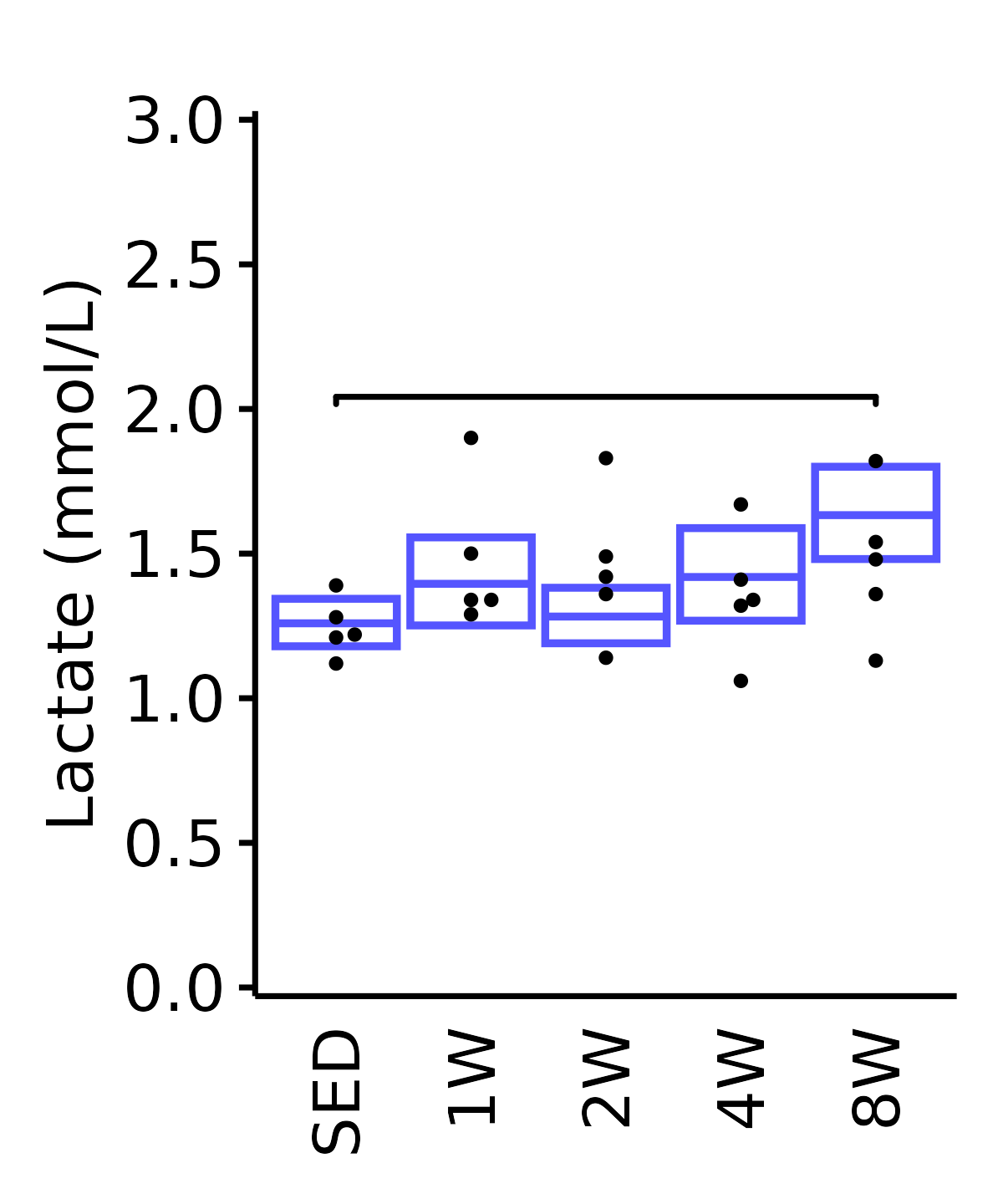
Leptin
Leptin (pg/mL).
6M Female
plot_baseline(ANALYTES,
response = "leptin",
conf = filter(conf_df, response == "Leptin"),
stats = filter(ANALYTES_STATS, response == "Leptin"),
sex = "Female", age = "6M", y_position = 3.1e4,
step_increase = 0.12) +
scale_y_continuous(name = "Leptin (pg/mL)",
labels = scales::label_scientific(),
breaks = seq(0, 7e4, 2e4),
limits = c(0, 7e4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(clip = "off", ylim = c(0, 7e4)) +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))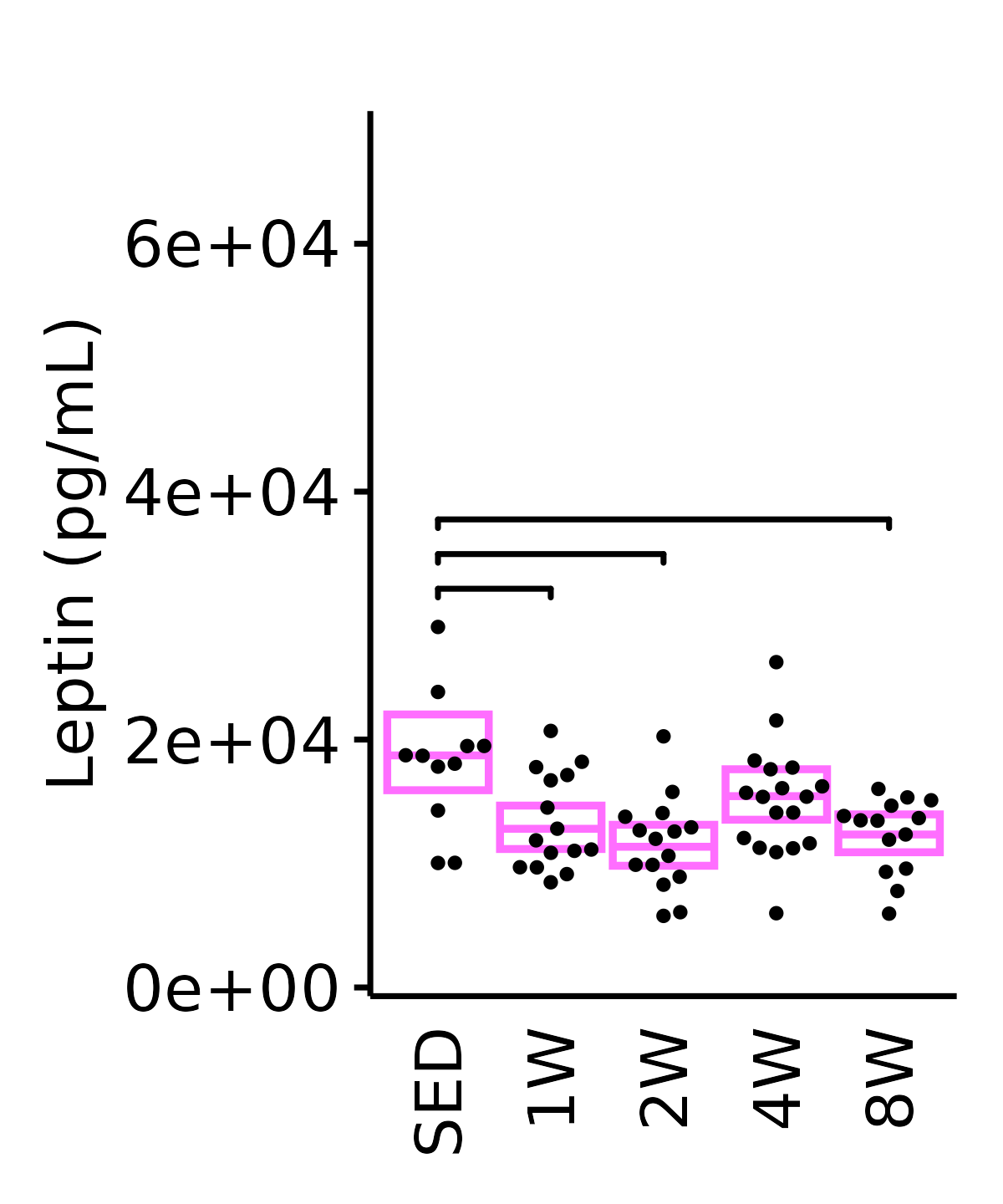
6M Male
plot_baseline(ANALYTES,
response = "leptin",
conf = filter(conf_df, response == "Leptin"),
stats = filter(ANALYTES_STATS, response == "Leptin"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "Leptin (pg/mL)",
labels = scales::label_scientific(),
breaks = seq(0, 7e4, 2e4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 7e4), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))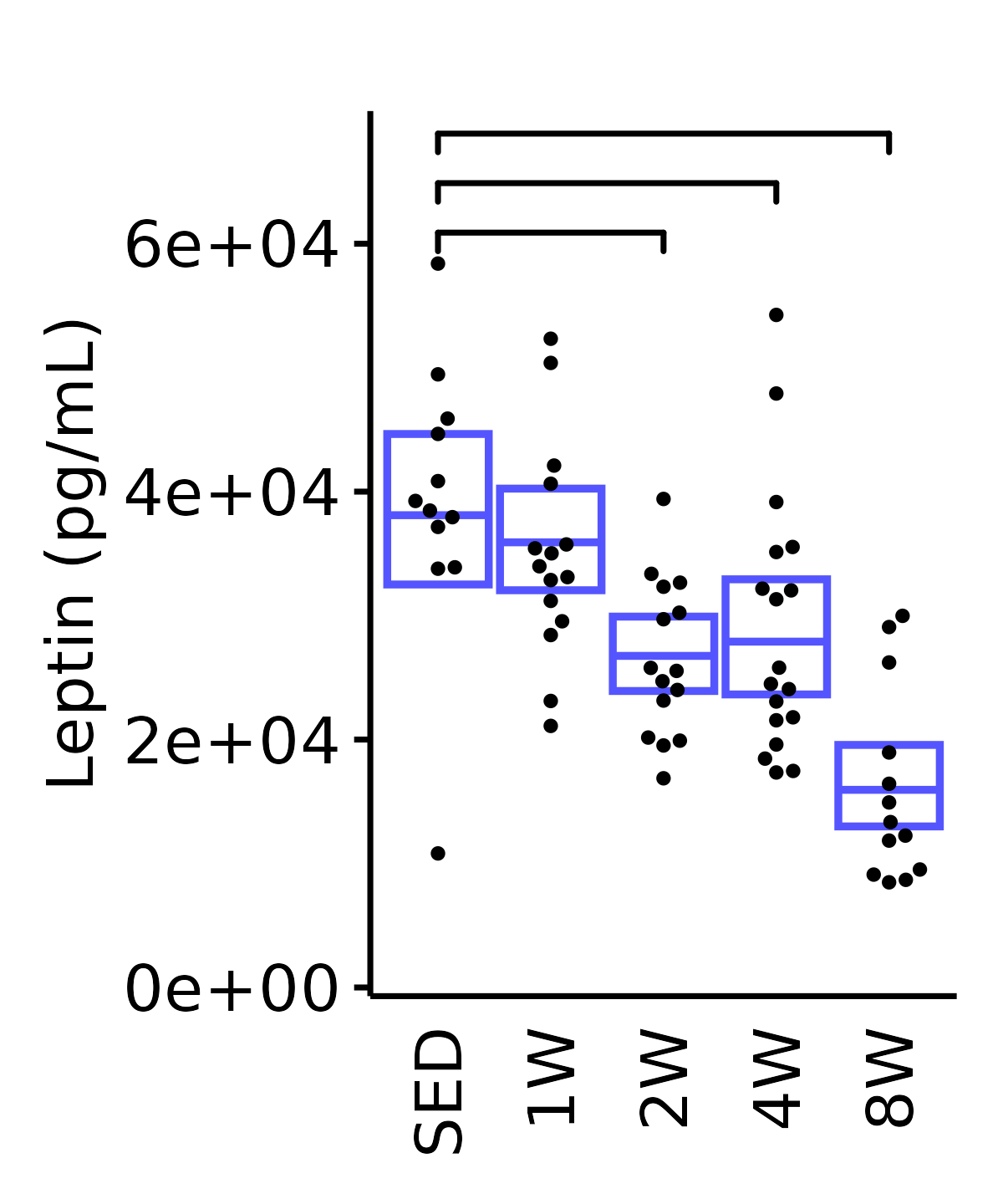
18M Female
plot_baseline(ANALYTES,
response = "leptin",
conf = filter(conf_df, response == "Leptin"),
stats = filter(ANALYTES_STATS, response == "Leptin"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "Leptin (pg/mL)",
labels = scales::label_scientific(),
breaks = seq(0, 7e4, 2e4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 7e4), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))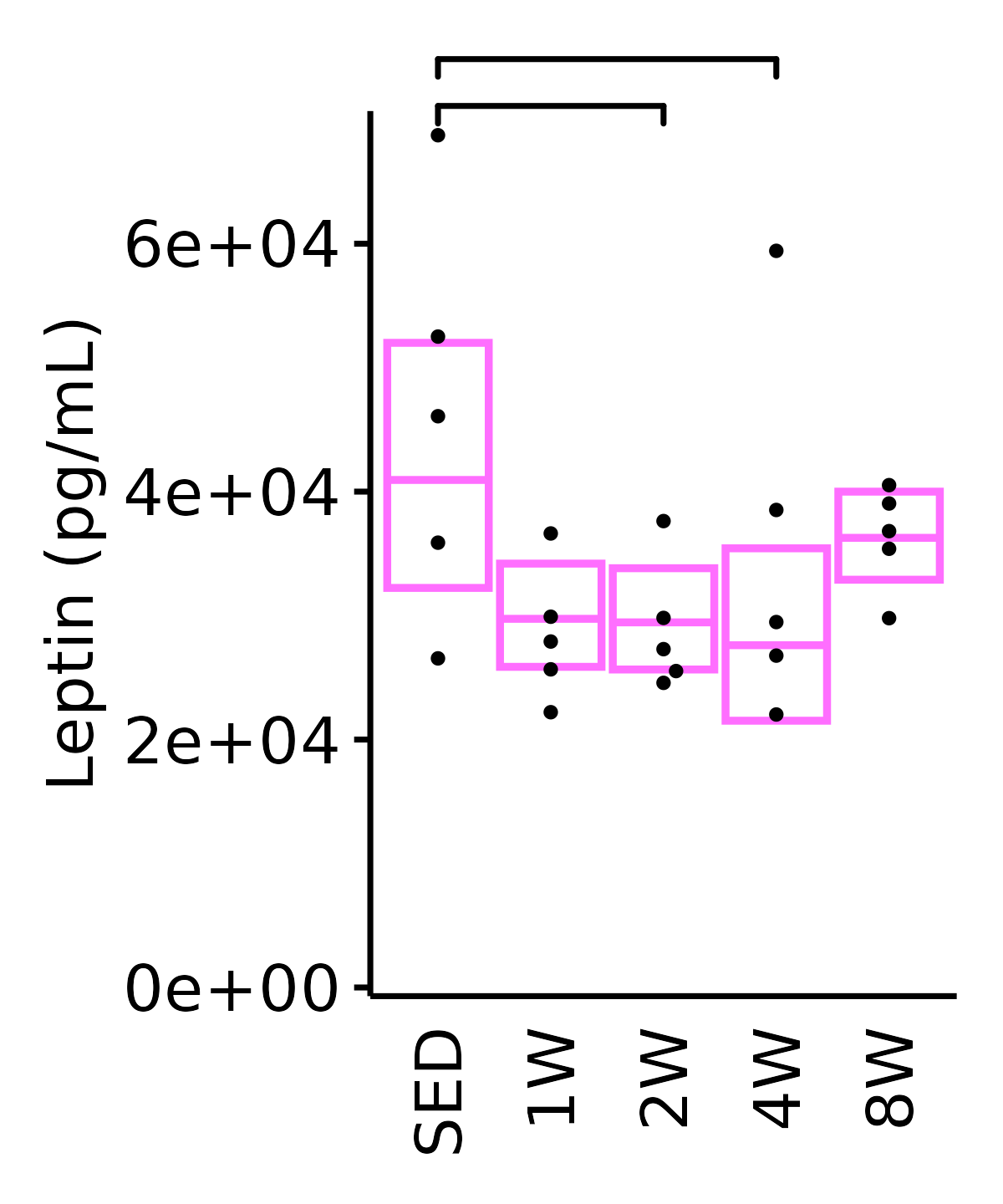
18M Male
plot_baseline(ANALYTES,
response = "leptin",
conf = filter(conf_df, response == "Leptin"),
stats = filter(ANALYTES_STATS, response == "Leptin"),
sex = "Male", age = "18M", y_position = 6e4) +
scale_y_continuous(name = "Leptin (pg/mL)",
labels = scales::label_scientific(),
breaks = seq(0, 7e4, 2e4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 7e4), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))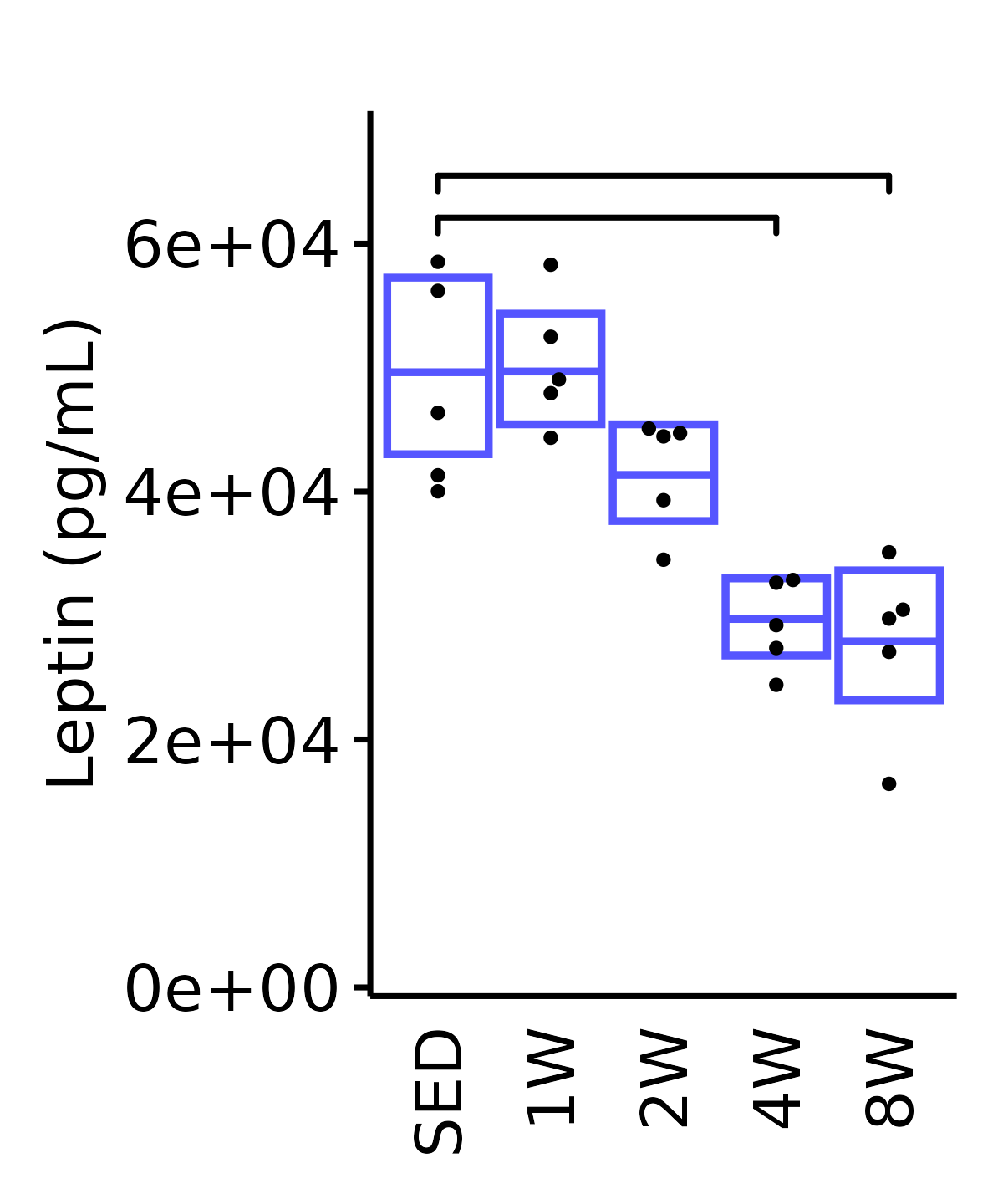
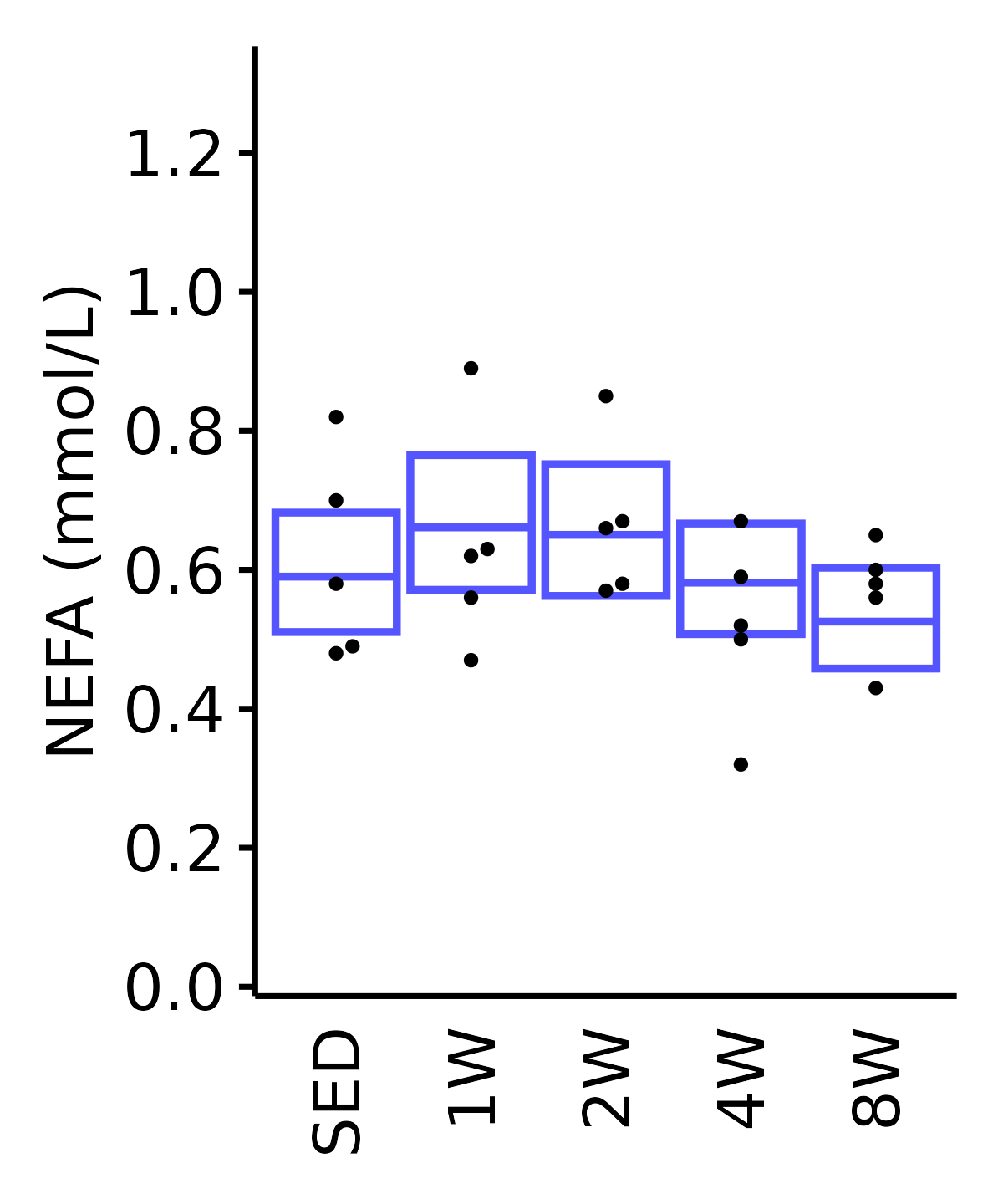
NEFA
NEFA (mmol/L).
6M Female
plot_baseline(ANALYTES,
response = "nefa",
conf = filter(conf_df, response == "NEFA"),
stats = filter(ANALYTES_STATS, response == "NEFA"),
sex = "Female", age = "6M", y_position = 1.25) +
scale_y_continuous(name = "NEFA (mmol/L)",
breaks = seq(0, 1.3, 0.2),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1.34), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))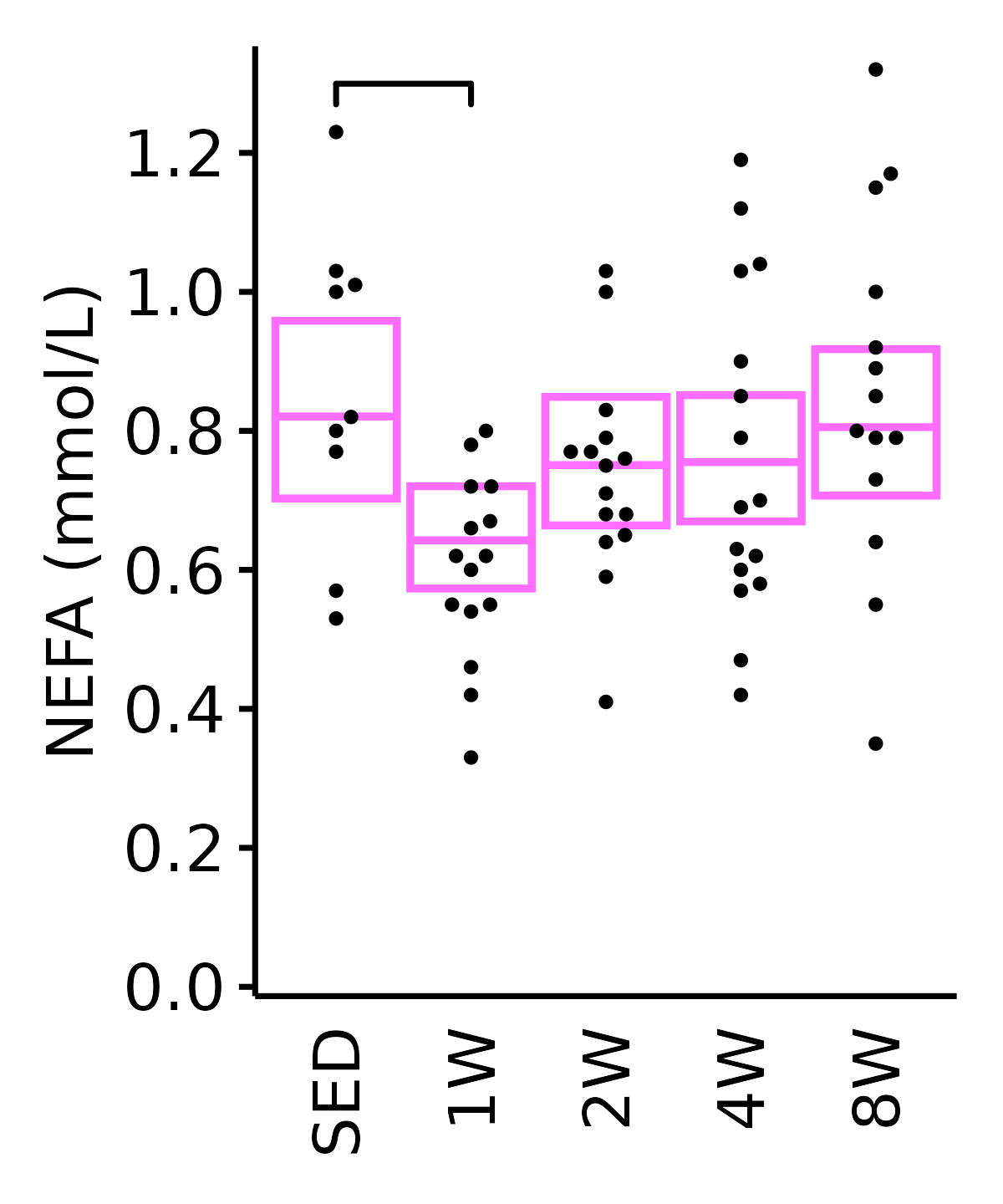
6M Male
plot_baseline(ANALYTES,
response = "nefa",
conf = filter(conf_df, response == "NEFA"),
stats = filter(ANALYTES_STATS, response == "NEFA"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "NEFA (mmol/L)",
breaks = seq(0, 1.3, 0.2),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1.34), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))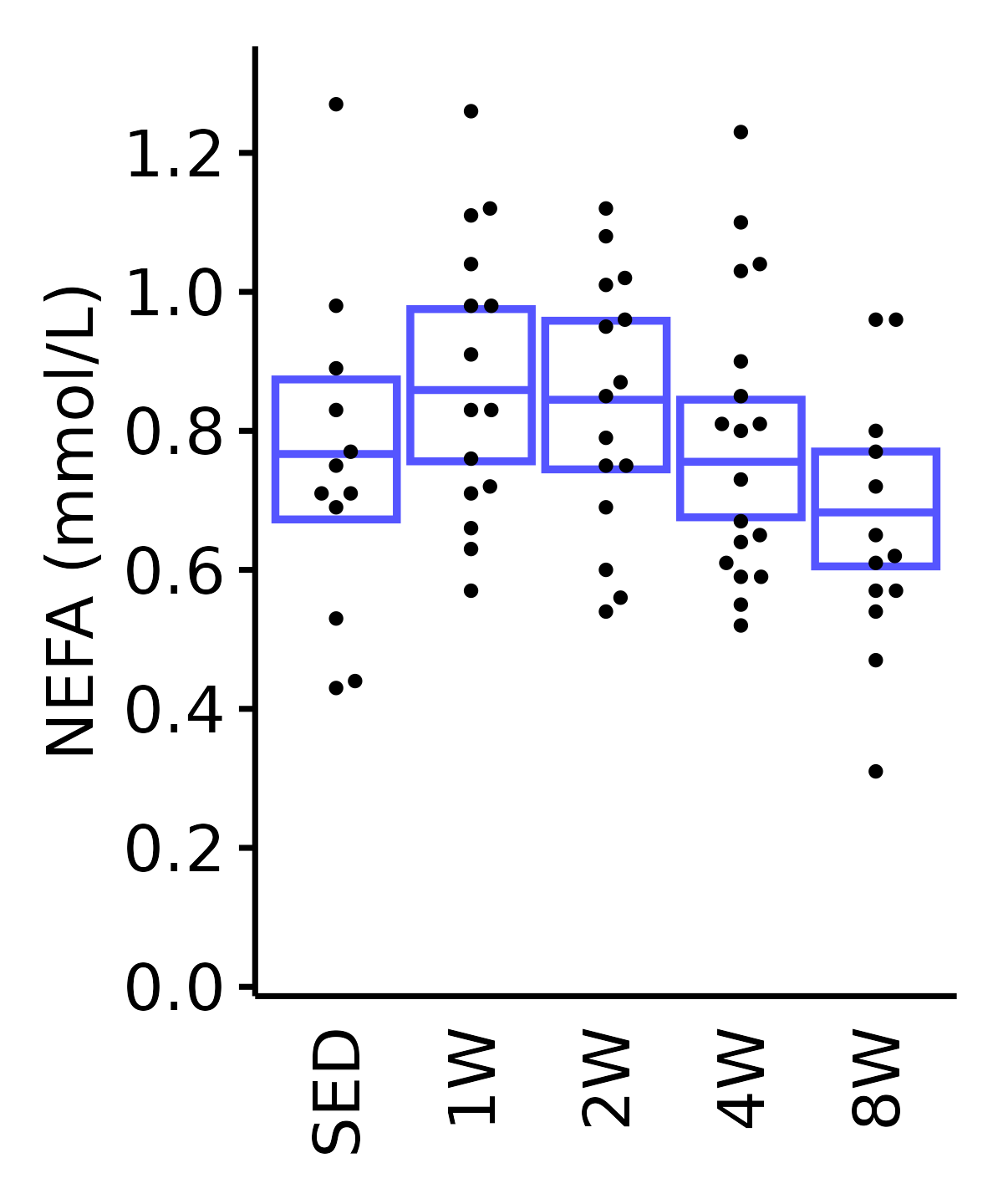
18M Female
plot_baseline(ANALYTES,
response = "nefa",
conf = filter(conf_df, response == "NEFA"),
stats = filter(ANALYTES_STATS, response == "NEFA"),
sex = "Female", age = "18M", y_position = 1.15) +
scale_y_continuous(name = "NEFA (mmol/L)",
breaks = seq(0, 1.3, 0.2),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1.34), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))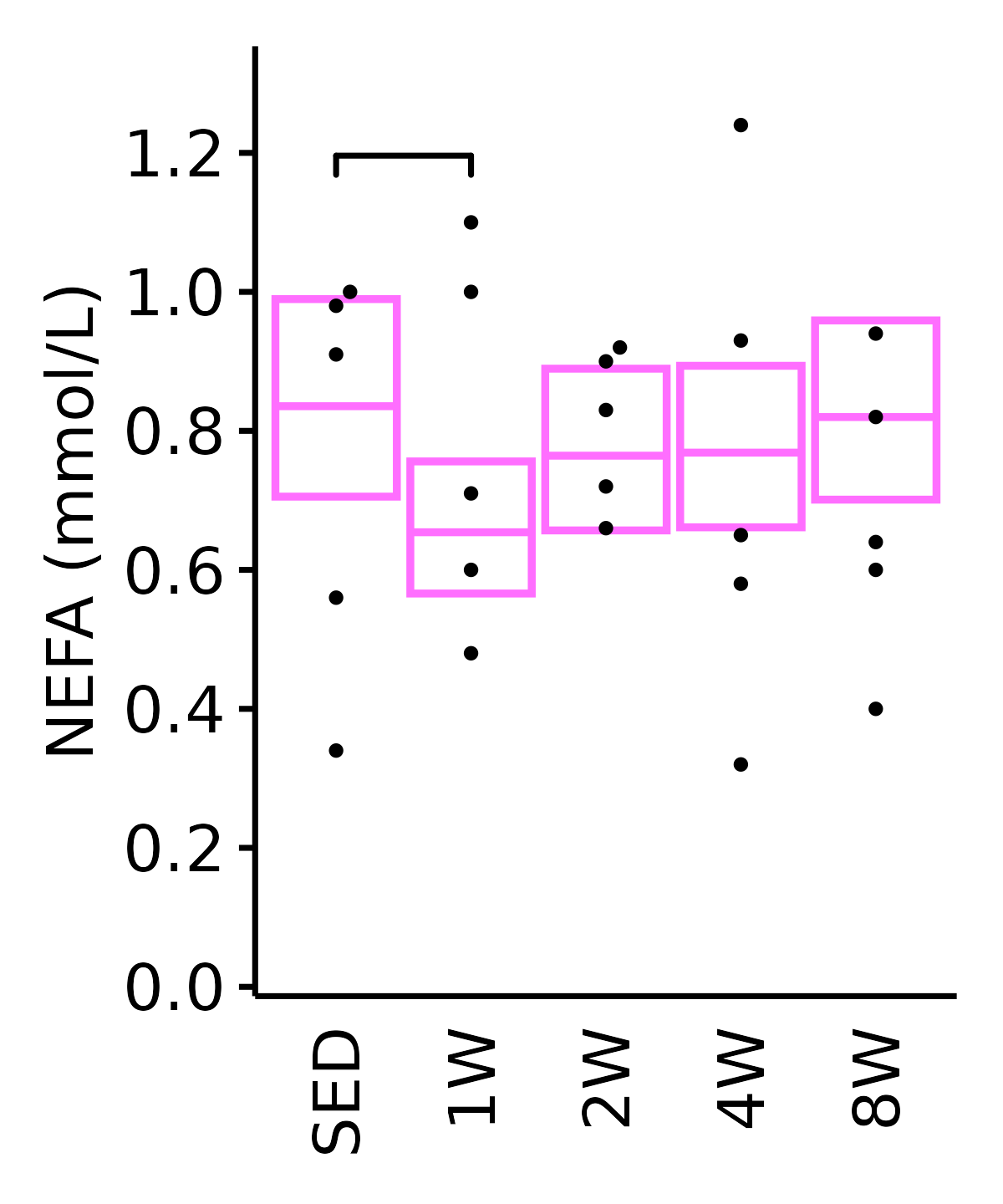
18M Male
plot_baseline(ANALYTES,
response = "nefa",
conf = filter(conf_df, response == "NEFA"),
stats = filter(ANALYTES_STATS, response == "NEFA"),
sex = "Male", age = "18M") +
scale_y_continuous(name = "NEFA (mmol/L)",
breaks = seq(0, 1.3, 0.2),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 1.34), clip = "off") +
theme(plot.margin = unit(c(5, 5, 5, 5), units = "pt"))