Statistical analysis of mean fiber area by muscle and fiber type
Tyler Sagendorf
01 May, 2024
Source:vignettes/FIBER_AREA_STATS.Rmd
FIBER_AREA_STATS.Rmd
# Required packages
library(MotrpacRatTrainingPhysiologyData)
library(ggplot2)
library(MASS)
library(dplyr)
library(tibble)
library(tidyr)
library(purrr)
library(emmeans)
library(nlme)
library(stats)
theme_set(theme_bw()) # base plot themePlots
We will look at plots of mean fiber area.
MG
# Plot points
FIBER_TYPES %>%
filter(muscle == "MG") %>%
ggplot(aes(x = group, y = fiber_area)) +
geom_point(na.rm = TRUE, alpha = 0.5,
position = position_jitter(width = 0.1, height = 0)) +
facet_grid(age + sex ~ type)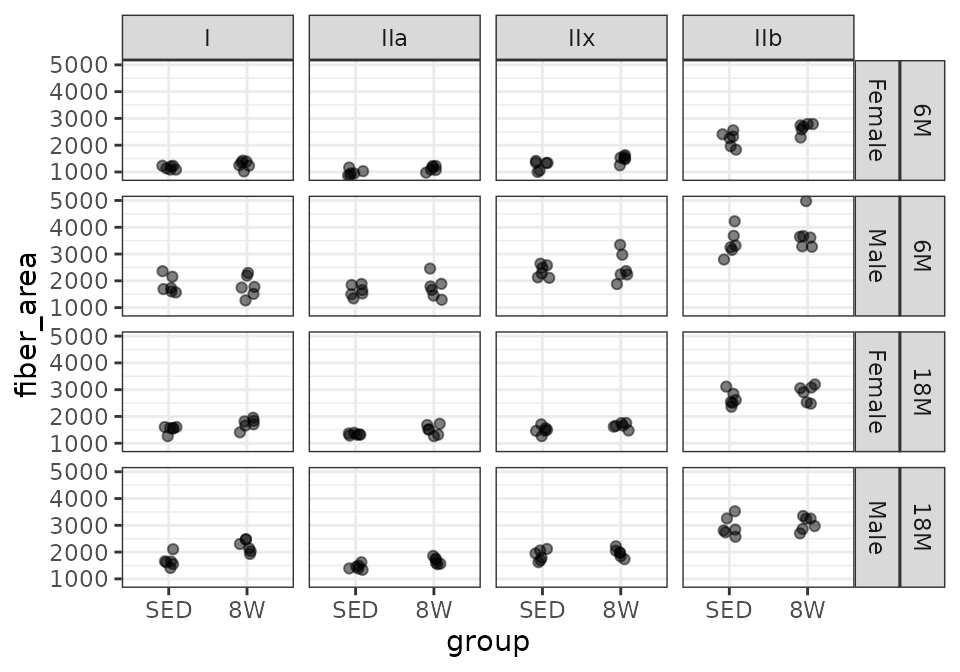
LG
# Plot points
FIBER_TYPES %>%
filter(muscle == "LG") %>%
ggplot(aes(x = group, y = fiber_area)) +
geom_point(na.rm = TRUE, alpha = 0.5,
position = position_jitter(width = 0.1, height = 0)) +
facet_grid(age + sex ~ type)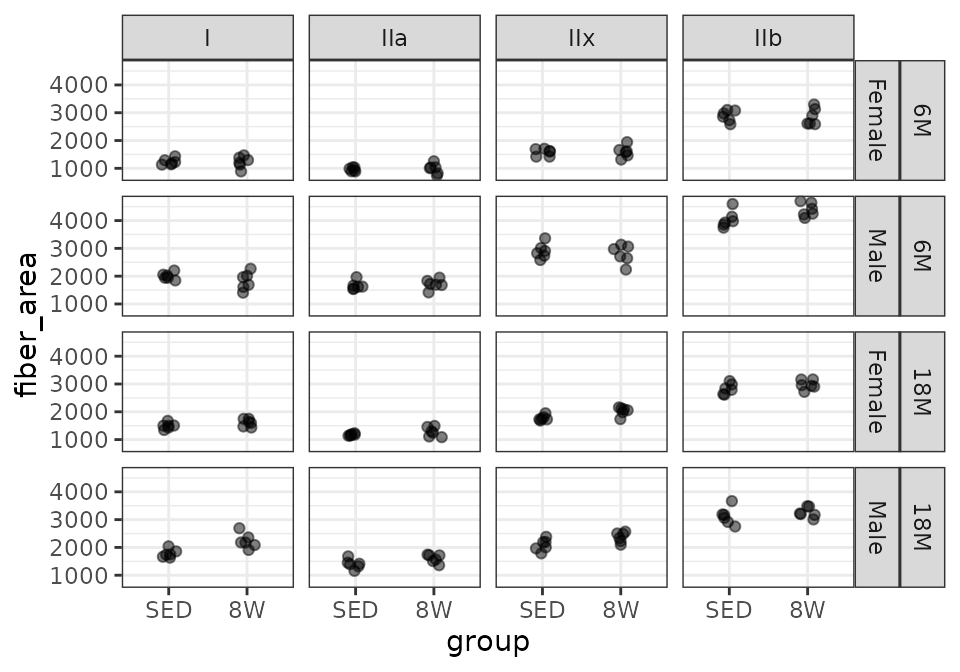
PL
# Plot points
FIBER_TYPES %>%
filter(muscle == "PL") %>%
ggplot(aes(x = group, y = fiber_area)) +
geom_point(na.rm = TRUE, alpha = 0.5,
position = position_jitter(width = 0.1, height = 0)) +
facet_grid(age + sex ~ type)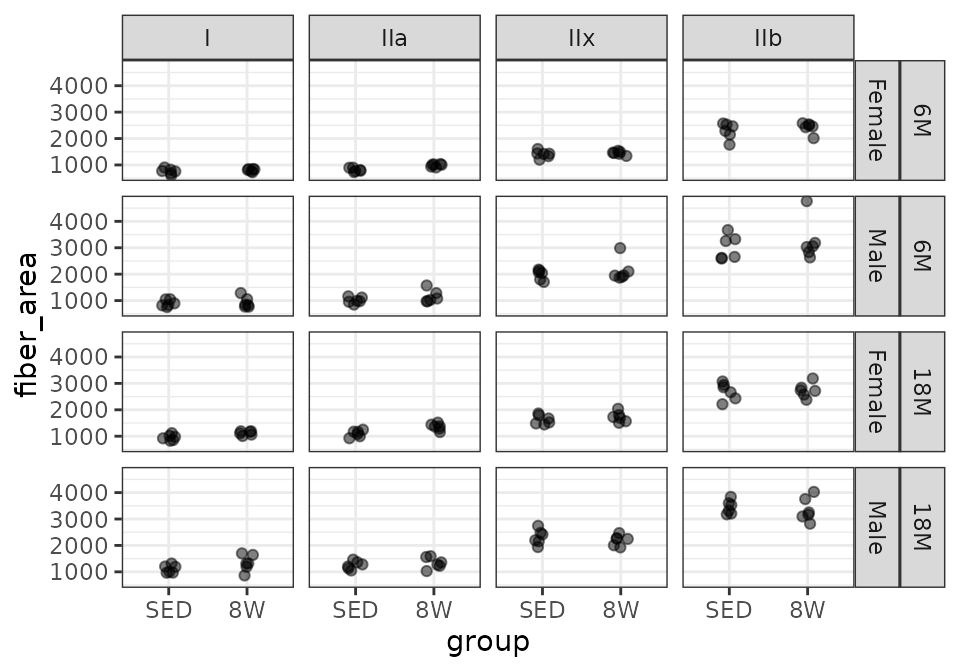
SOL
# Plot points
FIBER_TYPES %>%
filter(muscle == "SOL") %>%
ggplot(aes(x = group, y = fiber_area)) +
geom_point(na.rm = TRUE, alpha = 0.5,
position = position_jitter(width = 0.1, height = 0)) +
facet_grid(age + sex ~ type)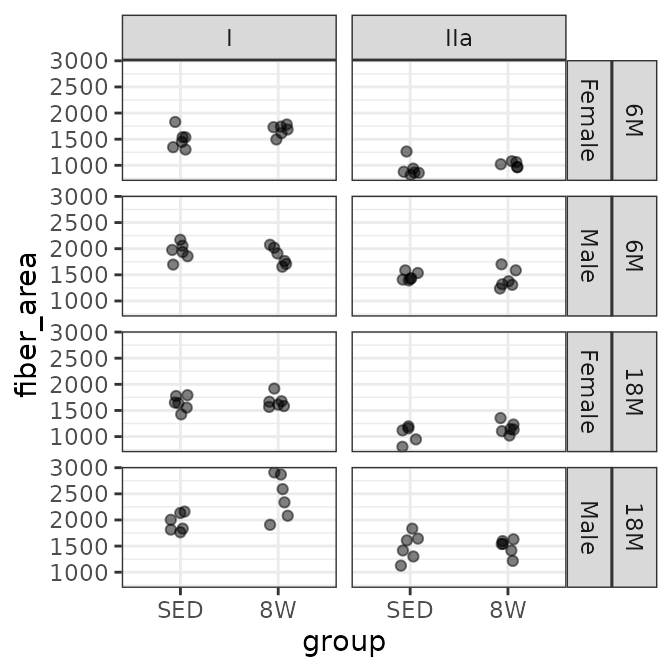
Regression Models
We will check the mean-variance relationship.
mv <- FIBER_TYPES %>%
group_by(sex, group, age, muscle, type) %>%
summarise(mn = mean(fiber_area, na.rm = TRUE),
vr = var(fiber_area, na.rm = TRUE))
fit.mv <- lm(log(vr) ~ log(mn), data = mv)
coef(fit.mv)
#> (Intercept) log(mn)
#> -4.451949 1.982954
plot(log(vr) ~ log(mn), data = mv, las = 1, pch = 19,
xlab = "log(group means)", ylab = "log(group variances)")
abline(coef(fit.mv), lwd = 2)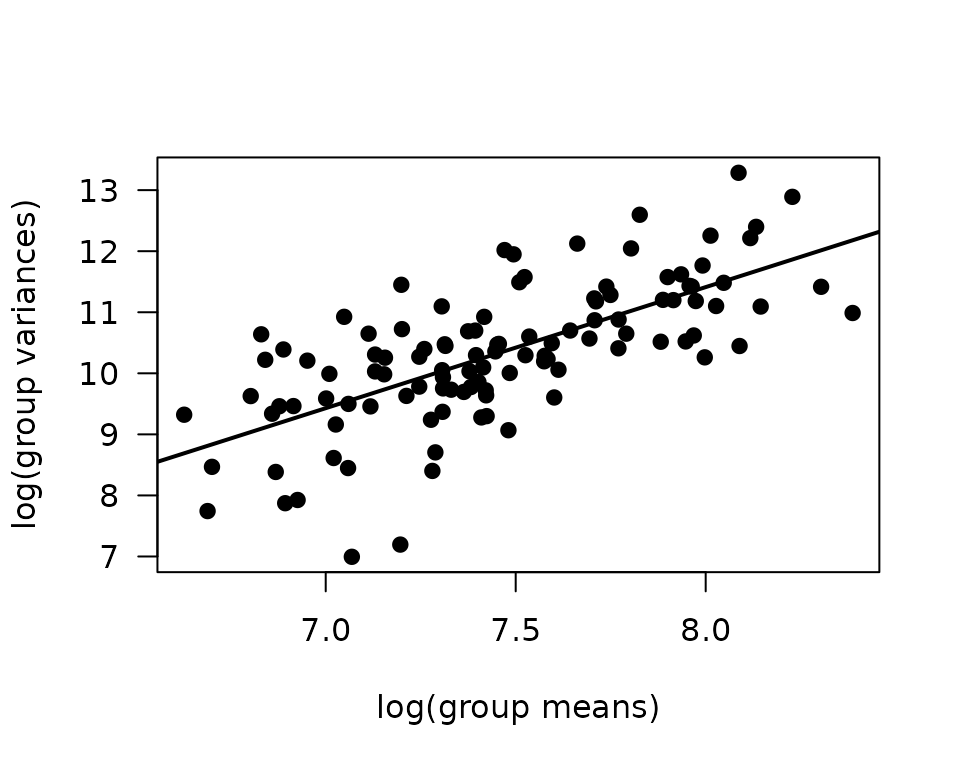
The slope of the line is close to 2, which suggests a gamma
distribution or log transformation may be appropriate. Since SOL only
consists of type I and IIa fibers, we can not include an interaction
between muscle and type, since it will be
inestimable for SOL type IIb and IIx. Instead, we will create a new
muscle_type variable by concatenating muscle
and type.
FIBER_TYPES <- mutate(FIBER_TYPES, muscle_type = paste0(muscle, ".", type))
fit.area <- lme(fixed = log(fiber_area) ~ age * sex * group * muscle_type,
random = ~ 1 | pid,
data = FIBER_TYPES,
na.action = na.exclude)We will check regression diagnostic plots and plots of residuals vs. predictor levels.
r <- resid(fit.area, scaled = TRUE, type = "pearson")
par(mfrow = c(2, 2))
# Standardized residuals vs. fitted plot
plot(x = fitted(fit.area), y = r,
ylab = "Standardized residuals", xlab = "Fitted values")
lines(loess.smooth(x = fitted(fit.area), y = r, degree = 2),
col = "red", lwd = 2)
# Scale-location plot
plot(x = fitted(fit.area), y = sqrt(abs(r)),
ylab = "sqrt(|standardized residuals|)", xlab = "Fitted values")
lines(loess.smooth(x = fitted(fit.area), y = sqrt(abs(r)), degree = 2),
col = "red", lwd = 2)
# Normal Q-Q plot
qqnorm(r); qqline(r, col = "red")
par(mfrow = c(1, 1))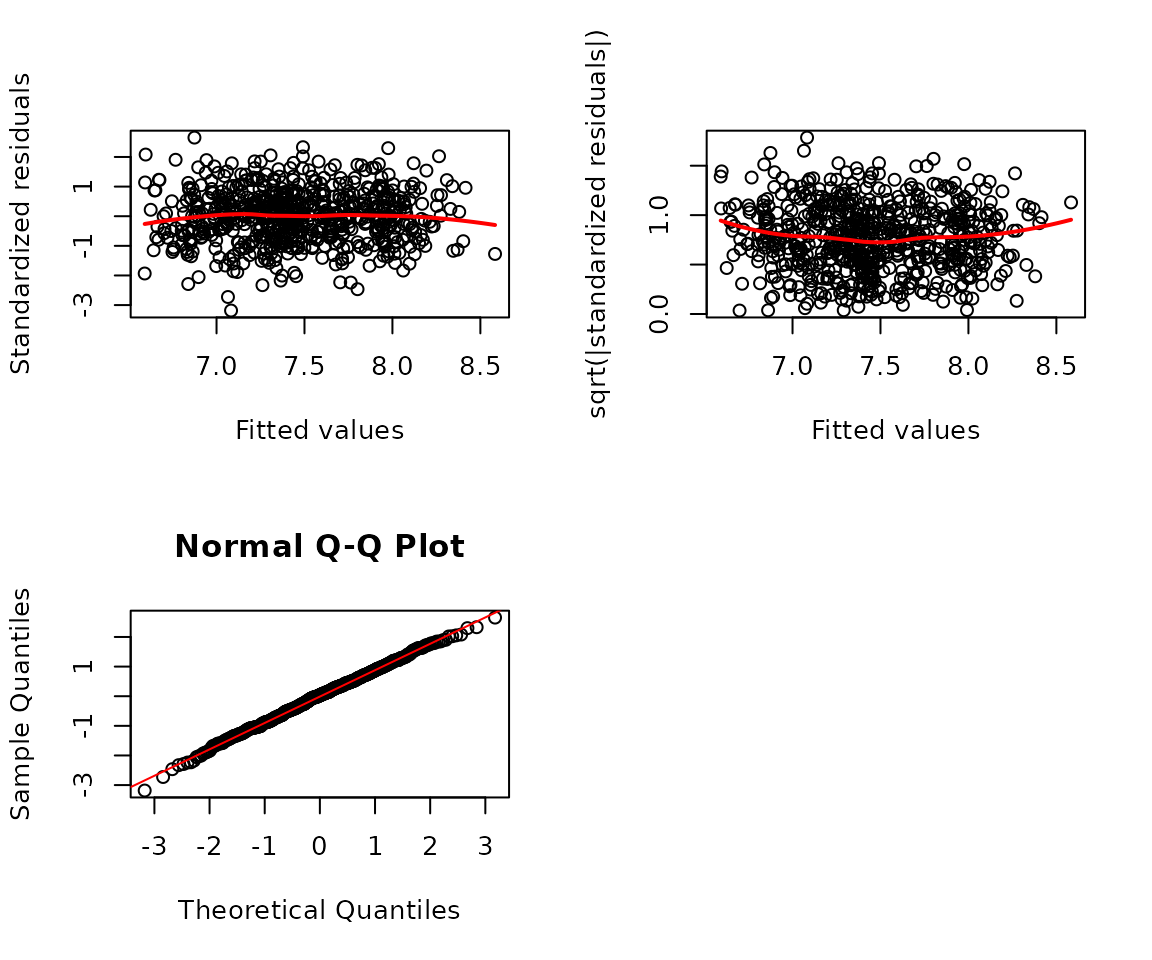
# Residuals vs. predictor levels
par(mfrow = c(2, 3), mar = c(5, 3, 0.5, 0.5))
boxplot(r ~ age, data = FIBER_TYPES, ylab = "")
boxplot(r ~ group, data = FIBER_TYPES, ylab = "")
boxplot(r ~ sex, data = FIBER_TYPES, ylab = "")
boxplot(r ~ muscle, data = FIBER_TYPES, ylab = "")
boxplot(r ~ type, data = FIBER_TYPES, ylab = "")
mtext("Standardized residuals", side = 2, line = -1, outer = TRUE)
par(mfrow = c(1, 1), mar = c(5, 4, 4, 2))
The diagnostic plots look fine.
anova(fit.area)
#> numDF denDF F-value p-value
#> (Intercept) 1 518 594342.4 <.0001
#> age 1 40 19.9 0.0001
#> sex 1 40 207.9 <.0001
#> group 1 40 15.2 0.0004
#> muscle_type 13 518 632.1 <.0001
#> age:sex 1 40 26.3 <.0001
#> age:group 1 40 1.5 0.2338
#> sex:group 1 40 0.5 0.5025
#> age:muscle_type 13 518 15.5 <.0001
#> sex:muscle_type 13 518 8.3 <.0001
#> group:muscle_type 13 518 1.8 0.0451
#> age:sex:group 1 40 0.8 0.3873
#> age:sex:muscle_type 13 518 7.7 <.0001
#> age:group:muscle_type 13 518 2.6 0.0017
#> sex:group:muscle_type 13 518 0.9 0.5899
#> age:sex:group:muscle_type 13 518 1.6 0.0821
VarCorr(fit.area)
#> pid = pdLogChol(1)
#> Variance StdDev
#> (Intercept) 0.003757709 0.06130015
#> Residual 0.010165279 0.10082301Hypothesis Testing
We will compare the 8W trained to the sedentary control group within each age, sex, and muscle using a two-sided t-test. We will then adjust p-values across fiber types within each age, sex, and muscle group using the Holm method.
# Estimated marginal means
FIBER_AREA_EMM <- emmeans(fit.area, specs = "group",
by = c("age", "sex", "muscle_type"),
type = "response", infer = TRUE)
# Extract model info
model_df <- data.frame(response = "Mean Fiber Area",
model = paste(deparse(fit.area[["call"]]),
collapse = "")) %>%
mutate(model = gsub("(?<=[\\s])\\s*|^\\s+|\\s+$", "", model, perl = TRUE),
model_type = "lme",
fixed = sub(".*fixed = ([^,]+),.*", "\\1", model),
random = "~1 | pid") %>%
dplyr::select(-model)
FIBER_AREA_STATS <- FIBER_AREA_EMM %>%
contrast(method = "trt.vs.ctrl") %>%
summary(infer = TRUE) %>%
as.data.frame() %>%
rename(any_of(c(lower.CL = "asymp.LCL",
upper.CL = "asymp.UCL"))) %>%
separate_wider_delim(cols = muscle_type, delim = ".",
names = c("muscle", "type")) %>%
mutate(muscle = factor(muscle, levels = c("LG", "MG", "PL", "SOL")),
type = factor(type, levels = c("I", "IIa", "IIx", "IIb"))) %>%
# Holm-adjust p-values across fiber types within each age, sex, and muscle
# combination
group_by(age, sex, muscle) %>%
mutate(p.adj = p.adjust(p.value, method = "holm"),
signif = p.adj < 0.05) %>%
ungroup() %>%
pivot_longer(cols = ratio,
names_to = "estimate_type",
values_to = "estimate",
values_drop_na = TRUE) %>%
pivot_longer(cols = contains(".ratio"),
names_to = "statistic_type",
values_to = "statistic",
values_drop_na = TRUE) %>%
mutate(response = "Mean Fiber Area",
statistic_type = sub("\\.ratio", "", statistic_type)) %>%
left_join(model_df, by = "response") %>%
# Reorder columns
dplyr::select(response, age, sex, muscle, type, contrast, estimate_type, null,
estimate, SE, lower.CL, upper.CL, statistic_type, statistic,
df, p.value, p.adj, signif, model_type, fixed, random,
everything()) %>%
arrange(response, age, sex, muscle, type) %>%
as.data.frame()See ?FIBER_AREA_STATS for details.
print.data.frame(head(FIBER_AREA_STATS))
#> response age sex muscle type contrast estimate_type null estimate
#> 1 Mean Fiber Area 6M Female LG I 8W / SED ratio 1 0.9858807
#> 2 Mean Fiber Area 6M Female LG IIa 8W / SED ratio 1 1.0077985
#> 3 Mean Fiber Area 6M Female LG IIx 8W / SED ratio 1 1.0069759
#> 4 Mean Fiber Area 6M Female LG IIb 8W / SED ratio 1 0.9865055
#> 5 Mean Fiber Area 6M Female MG I 8W / SED ratio 1 1.0947372
#> 6 Mean Fiber Area 6M Female MG IIa 8W / SED ratio 1 1.1627512
#> SE lower.CL upper.CL statistic_type statistic df p.value
#> 1 0.06716298 0.8590696 1.131411 t -0.2087328 40 0.8357162
#> 2 0.06865613 0.8781682 1.156564 t 0.1140301 40 0.9097843
#> 3 0.06860009 0.8774514 1.155620 t 0.1020439 40 0.9192317
#> 4 0.06720554 0.8596140 1.132128 t -0.1994340 40 0.8429336
#> 5 0.07457881 0.9539241 1.256336 t 1.3286536 40 0.1914926
#> 6 0.07921226 1.0131897 1.334390 t 2.2134207 40 0.0326355
#> p.adj signif model_type
#> 1 1.00000000 FALSE lme
#> 2 1.00000000 FALSE lme
#> 3 1.00000000 FALSE lme
#> 4 1.00000000 FALSE lme
#> 5 0.19149264 FALSE lme
#> 6 0.06527099 FALSE lme
#> fixed random
#> 1 log(fiber_area) ~ age * sex * group * muscle_type ~1 | pid
#> 2 log(fiber_area) ~ age * sex * group * muscle_type ~1 | pid
#> 3 log(fiber_area) ~ age * sex * group * muscle_type ~1 | pid
#> 4 log(fiber_area) ~ age * sex * group * muscle_type ~1 | pid
#> 5 log(fiber_area) ~ age * sex * group * muscle_type ~1 | pid
#> 6 log(fiber_area) ~ age * sex * group * muscle_type ~1 | pidSession Info
sessionInfo()
#> R version 4.4.0 (2024-04-24)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] nlme_3.1-164
#> [2] emmeans_1.10.1
#> [3] purrr_1.0.2
#> [4] tidyr_1.3.1
#> [5] tibble_3.2.1
#> [6] dplyr_1.1.4
#> [7] MASS_7.3-60.2
#> [8] ggplot2_3.5.1
#> [9] MotrpacRatTrainingPhysiologyData_2.0.0
#>
#> loaded via a namespace (and not attached):
#> [1] sass_0.4.9 utf8_1.2.4 generics_0.1.3 rstatix_0.7.2
#> [5] stringi_1.8.3 lattice_0.22-6 digest_0.6.35 magrittr_2.0.3
#> [9] estimability_1.5 evaluate_0.23 grid_4.4.0 mvtnorm_1.2-4
#> [13] fastmap_1.1.1 jsonlite_1.8.8 backports_1.4.1 fansi_1.0.6
#> [17] scales_1.3.0 textshaping_0.3.7 jquerylib_0.1.4 abind_1.4-5
#> [21] cli_3.6.2 rlang_1.1.3 munsell_0.5.1 withr_3.0.0
#> [25] cachem_1.0.8 yaml_2.3.8 ggbeeswarm_0.7.2 tools_4.4.0
#> [29] memoise_2.0.1 ggsignif_0.6.4 colorspace_2.1-0 ggpubr_0.6.0
#> [33] broom_1.0.5 vctrs_0.6.5 R6_2.5.1 lifecycle_1.0.4
#> [37] stringr_1.5.1 fs_1.6.4 car_3.1-2 htmlwidgets_1.6.4
#> [41] vipor_0.4.7 ragg_1.3.0 pkgconfig_2.0.3 beeswarm_0.4.0
#> [45] desc_1.4.3 pkgdown_2.0.9 pillar_1.9.0 bslib_0.7.0
#> [49] gtable_0.3.5 glue_1.7.0 systemfonts_1.0.6 highr_0.10
#> [53] xfun_0.43 tidyselect_1.2.1 knitr_1.46 farver_2.1.1
#> [57] htmltools_0.5.8.1 labeling_0.4.3 rmarkdown_2.26 carData_3.0-5
#> [61] compiler_4.4.0