Plots of weekly body mass and blood lactate
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/weekly_measures_plots.Rmd
weekly_measures_plots.Rmd
# Required packages
library(MotrpacRatTrainingPhysiologyData)
library(dplyr)
library(ggplot2)
library(tidyr)
library(emmeans)
library(latex2exp)Weekly Body Mass
Body mass (g) was measured at the start of each week prior to training.
6M Female
WEEKLY_BODY_MASS_EMM %>%
summary(type = "response", infer = TRUE) %>%
as.data.frame() %>%
dplyr::rename(week = week,
response_mean = response) %>%
filter(age == "6M", sex == "Female") %>%
ggplot(aes(x = week, y = response_mean)) +
geom_crossbar(aes(ymin = lower.CL, ymax = upper.CL),
fatten = 1, color = "#ff6eff",
width = 0.7, linewidth = 0.4) +
facet_grid(~ group) +
scale_y_continuous(name = "Body Mass (g)",
expand = expansion(mult = 5e-3),
breaks = seq(180, 250, 20)) +
coord_cartesian(ylim = c(175, 255), clip = "off") +
labs(x = "Week") +
theme_bw(base_size = 7) +
theme(text = element_text(color = "black"),
axis.text = element_text(color = "black", size = 7),
axis.title = element_text(color = "black", size = 7),
axis.ticks = element_line(color = "black"),
axis.ticks.x = element_blank(),
strip.text = element_text(color = "black", size = 7),
strip.background = element_blank(),
panel.spacing = unit(-1, "pt"),
panel.grid = element_blank())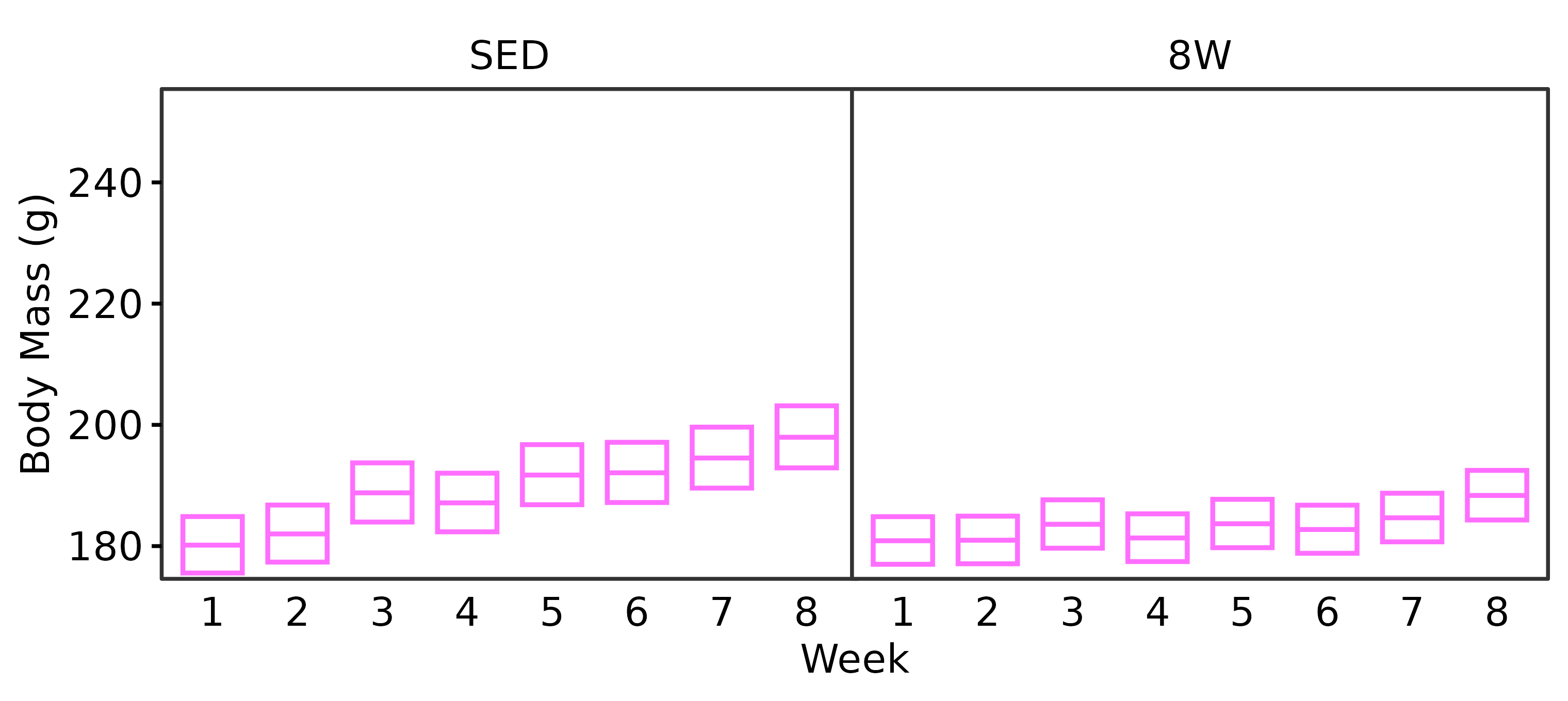
6M Male
WEEKLY_BODY_MASS_EMM %>%
summary(type = "response", infer = TRUE) %>%
as.data.frame() %>%
filter(age == "6M", sex == "Male") %>%
dplyr::rename(week = week,
response_mean = response) %>%
ggplot(aes(x = week, y = response_mean)) +
geom_crossbar(aes(ymin = lower.CL, ymax = upper.CL),
fatten = 1, color = "#5555ff",
width = 0.7, linewidth = 0.4) +
facet_grid(~ group) +
scale_y_continuous(name = "Body Mass (g)",
breaks = seq(300, 440, 40),
expand = expansion(mult = 5e-3)) +
coord_cartesian(ylim = c(300, 450), clip = "off") +
labs(x = "Week") +
theme_bw(base_size = 7) +
theme(text = element_text(color = "black"),
axis.text = element_text(color = "black", size = 7),
axis.title = element_text(color = "black", size = 7),
axis.ticks = element_line(color = "black"),
axis.ticks.x = element_blank(),
strip.text = element_text(color = "black", size = 7),
strip.background = element_blank(),
panel.spacing = unit(-1, "pt"),
panel.grid = element_blank())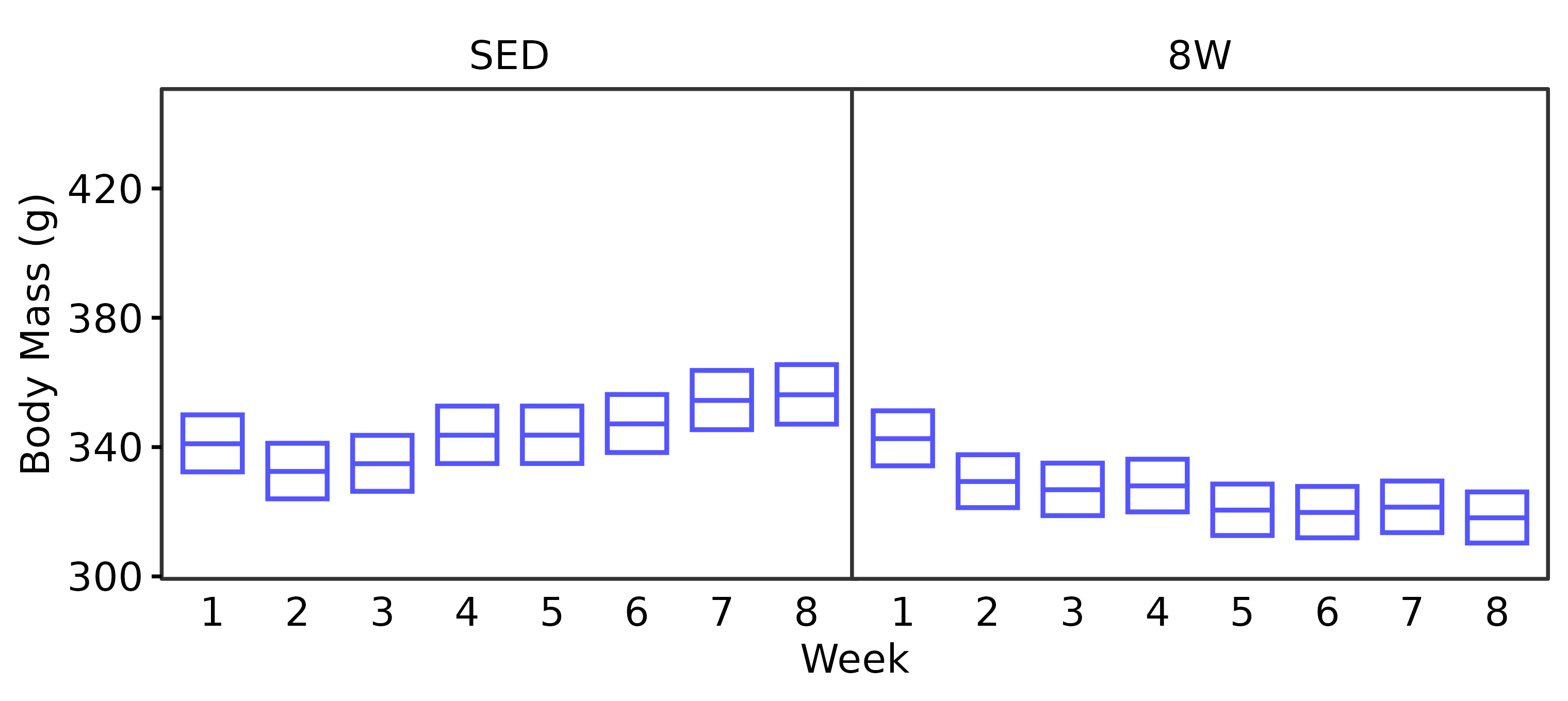
18M Female
WEEKLY_BODY_MASS_EMM %>%
summary(type = "response", infer = TRUE) %>%
as.data.frame() %>%
filter(age == "18M", sex == "Female") %>%
dplyr::rename(week = week,
response_mean = response) %>%
ggplot(aes(x = week, y = response_mean)) +
geom_crossbar(aes(ymin = lower.CL, ymax = upper.CL),
fatten = 1, color = "#ff6eff",
width = 0.7, linewidth = 0.4) +
facet_grid(~ group) +
scale_y_continuous(name = "Body Mass (g)",
expand = expansion(mult = 5e-3),
breaks = seq(180, 250, 20)) +
coord_cartesian(ylim = c(175, 255), clip = "off") +
labs(x = "Week") +
theme_bw(base_size = 7) +
theme(text = element_text(color = "black"),
axis.text = element_text(color = "black", size = 7),
axis.title = element_text(color = "black", size = 7),
axis.ticks = element_line(color = "black"),
axis.ticks.x = element_blank(),
strip.text = element_text(color = "black", size = 7),
strip.background = element_blank(),
panel.spacing = unit(-1, "pt"),
panel.grid = element_blank())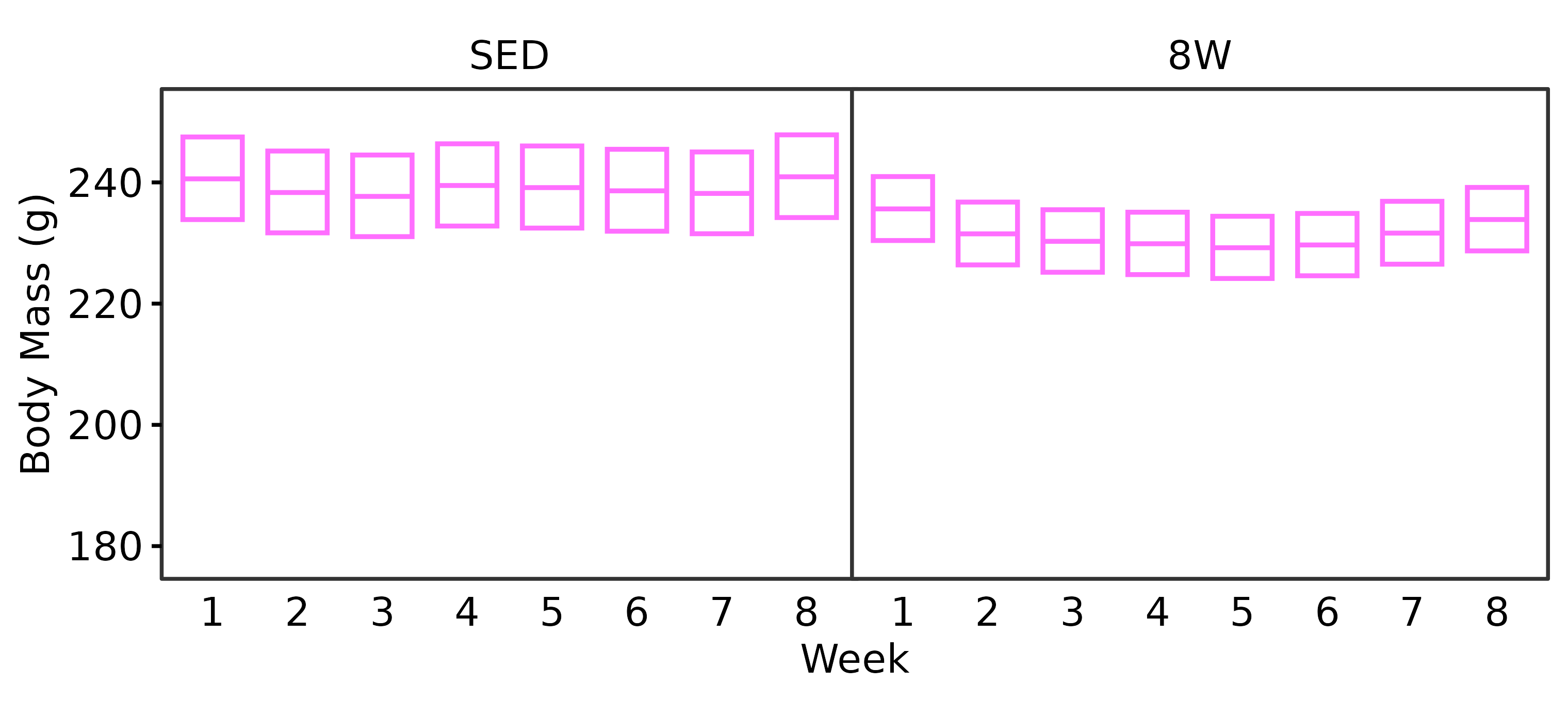
18M Male
WEEKLY_BODY_MASS_EMM %>%
summary(type = "response", infer = TRUE) %>%
as.data.frame() %>%
dplyr::rename(week = week,
response_mean = response) %>%
filter(age == "18M", sex == "Male") %>%
ggplot(aes(x = week, y = response_mean)) +
geom_crossbar(aes(ymin = lower.CL, ymax = upper.CL),
fatten = 1, color = "#5555ff",
width = 0.7, linewidth = 0.4) +
facet_grid(~ group) +
scale_y_continuous(name = "Body Mass (g)",
breaks = seq(300, 440, 40),
expand = expansion(mult = 5e-3)) +
coord_cartesian(ylim = c(300, 450), clip = "off") +
labs(x = "Week") +
theme_bw(base_size = 7) +
theme(text = element_text(color = "black"),
axis.text = element_text(color = "black", size = 7),
axis.title = element_text(color = "black", size = 7),
axis.ticks = element_line(color = "black"),
axis.ticks.x = element_blank(),
strip.text = element_text(color = "black", size = 7),
strip.background = element_blank(),
panel.spacing = unit(-1, "pt"),
panel.grid = element_blank())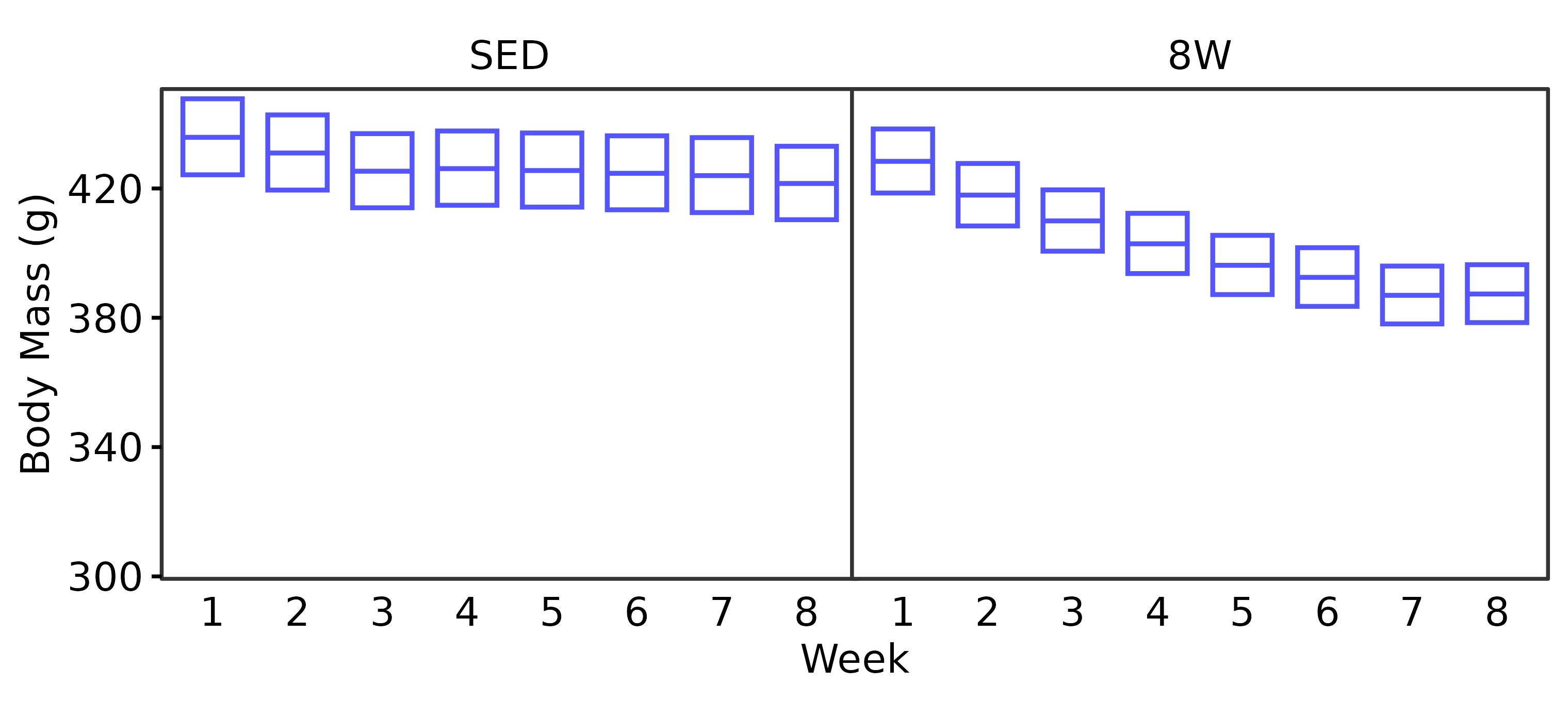
Plot Analysis Results
WEEKLY_BODY_MASS_STATS %>%
mutate(age = factor(age, levels = c("6M", "18M"),
labels = c("Adult", "Aged"))) %>%
ggplot(aes(x = as.numeric(week), y = estimate, color = sex)) +
geom_line(aes(x = as.numeric(week), group = sex),
position = position_dodge(width = 0.5)) +
geom_errorbar(aes(ymin = lower.CL, ymax = upper.CL,
color = sex), width = 0.6, linewidth = 0.5,
position = position_dodge(width = 0.5)) +
geom_point(aes(group = sex, shape = signif),
position = position_dodge(width = 0.5),
size = 1.5, fill = "white") +
facet_grid(~ age) +
geom_hline(yintercept = 1, lty = "dashed") +
scale_color_manual(values = c("#ff6eff", "#556eff")) +
scale_shape_manual(name = "Benjamini-Yekutieli-adjusted P-value:",
values = c(21, 16),
labels = c(latex2exp::TeX("$\\geq 0.05$"), "< 0.05")) +
guides(color = guide_none()) +
scale_y_continuous(limits = c(0.85, 1.05),
breaks = seq(0.85, 1.05, 0.05),
expand = expansion(mult = 5e-3)) +
labs(x = "Week", y = "8W / SED Mean Ratio") +
theme_bw(base_size = 7) +
theme(panel.border = element_rect(color = NA),
panel.grid = element_blank(),
axis.line = element_line(color = "black"),
text = element_text(color = "black"),
legend.position = "bottom",
legend.direction = "horizontal",
legend.margin = margin(t = -2, b = 0),
legend.key.size = unit(7, "pt"),
legend.text = element_text(color = "black", size = 7),
legend.title = element_text(color = "black", size = 7),
axis.text = element_text(color = "black", size = 7),
axis.title = element_text(color = "black", size = 7),
axis.ticks = element_line(color = "black"),
strip.text = element_text(color = "black", size = 7),
strip.background = element_blank())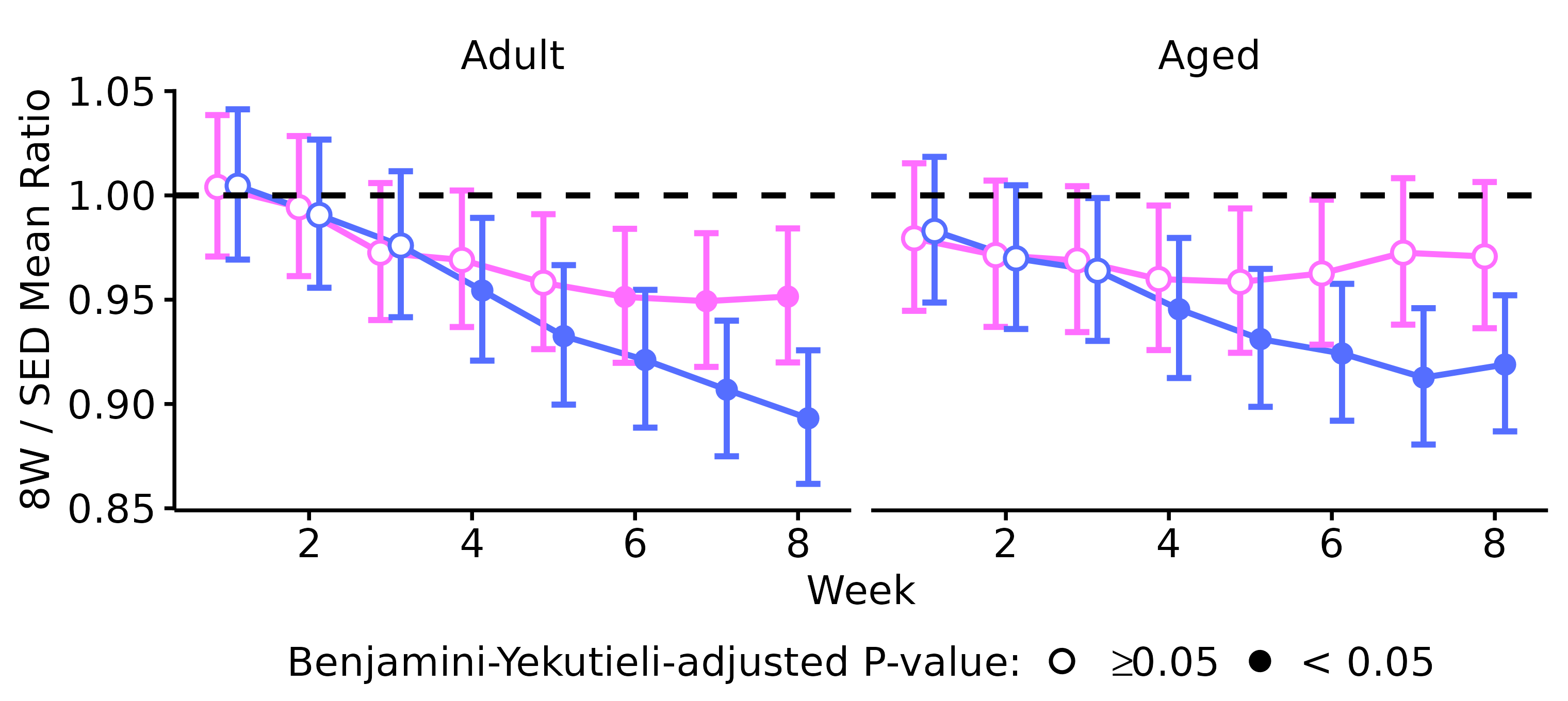
ggsave(filename = file.path("..", "..", "plots", "weekly_body_mass_tests.pdf"),
height = 2.4, width = 3.8, family = "ArialMT")Both sexes lose a significant amount of body mass after 3 weeks of exercise. While males continue to lose body mass throughout the protocol, female body mass appears to stabilize starting after 4 weeks of exercise. That is, they experience an initial decrease in body mass that is maintained with training.
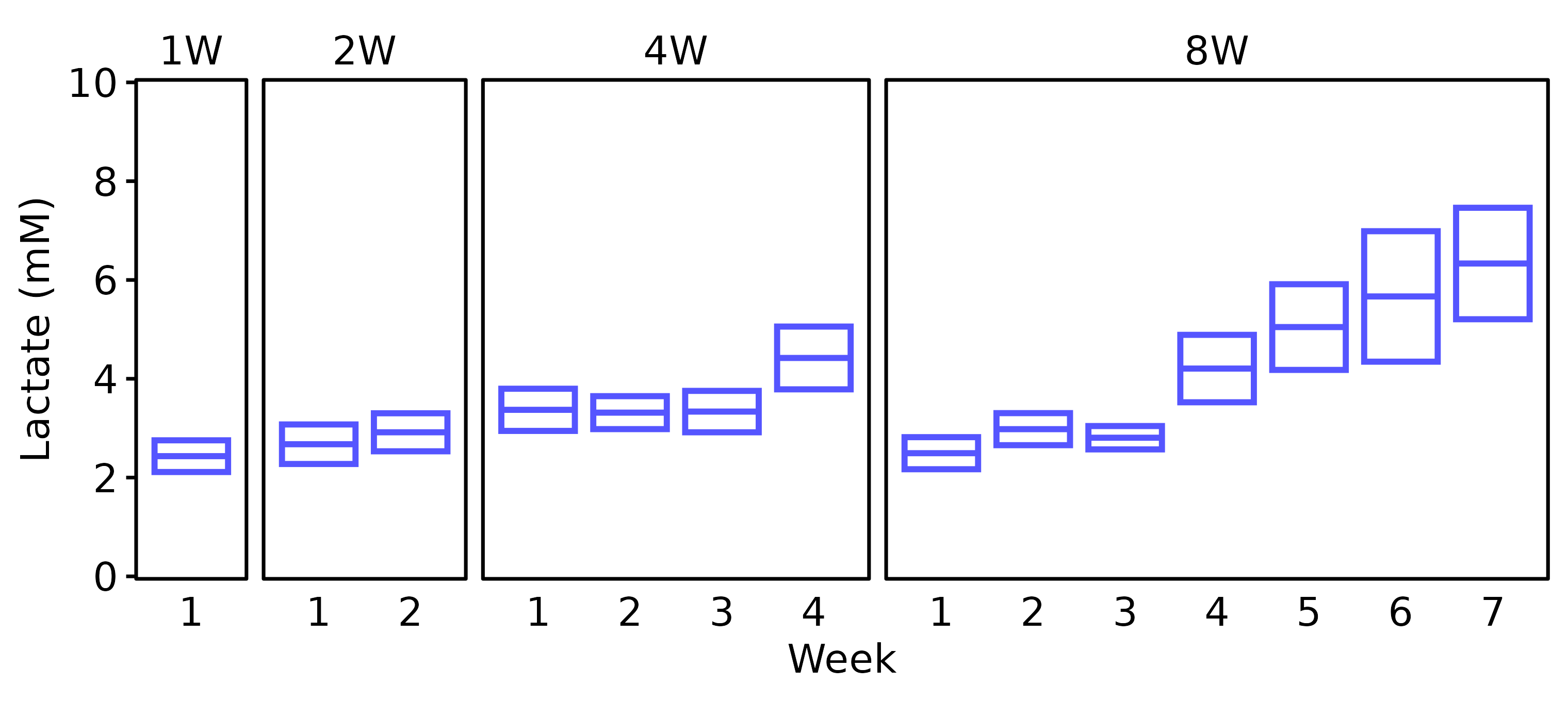
Lactate
Blood lactate was recorded at the start and end of each week at the end of the training bout. For the SED group, blood lactate was only recorded in 18M males, so we will not plot SED values.
lact_df <- WEEKLY_MEASURES %>%
dplyr::select(pid, age, sex, group, week, week_time, lactate) %>%
distinct() %>%
filter(!is.na(lactate)) %>%
filter(group != "SED") %>%
mutate(week_time = factor(week_time, levels = c("start", "end"),
labels = c("Start", "End")),
week = factor(week, levels = 1:8, labels = paste0("", 1:8))) %>%
droplevels.data.frame() %>%
group_by(age, sex, group, week, week_time) %>%
mutate(n = n()) %>%
filter(week_time == "Start")
# Reusable lactate plot code
plot_lactate <- function(x) {
ggplot() +
stat_summary(geom = "crossbar", fun.data = mean_cl_normal,
data = filter(x, n >= 5),
mapping = aes(x = week, y = lactate, color = sex),
width = 0.8, linewidth = 0.5, fatten = 1) +
geom_point(aes(x = week, y = lactate, color = sex),
data = filter(x, n < 5),
position = ggbeeswarm::position_beeswarm(cex = 2,
dodge.width = 0.7),
shape = 16, size = 0.5) +
facet_grid(~ group, space = "free", scales = "free") +
scale_color_manual(values = c("#ff6eff", "#5555ff"),
breaks = c("Female", "Male")) +
labs(x = "Week") +
theme_bw(base_size = 7) +
theme(text = element_text(size = 7, color = "black"),
line = element_line(linewidth = 0.3, color = "black"),
axis.ticks = element_line(linewidth = 0.3, color = "black"),
panel.grid = element_blank(),
panel.border = element_rect(color = "black", fill = NA,
linewidth = 0.3),
axis.ticks.x = element_blank(),
axis.text = element_text(size = 7, color = "black"),
axis.title = element_text(size = 7, margin = margin(),
color = "black"),
strip.background = element_blank(),
strip.text = element_text(size = 7, color = "black",
margin = margin(b = 2, t = 2)),
panel.spacing = unit(3, "pt"),
legend.position = "none"
)
}6M Female
# 6M Female
lact_df %>%
filter(age == "6M", sex == "Female") %>%
plot_lactate() +
scale_y_continuous(name = "Lactate (mM)",
expand = expansion(mult = 5e-3),
breaks = seq(0, 10, 2)) +
coord_cartesian(ylim = c(0, 10), clip = "off")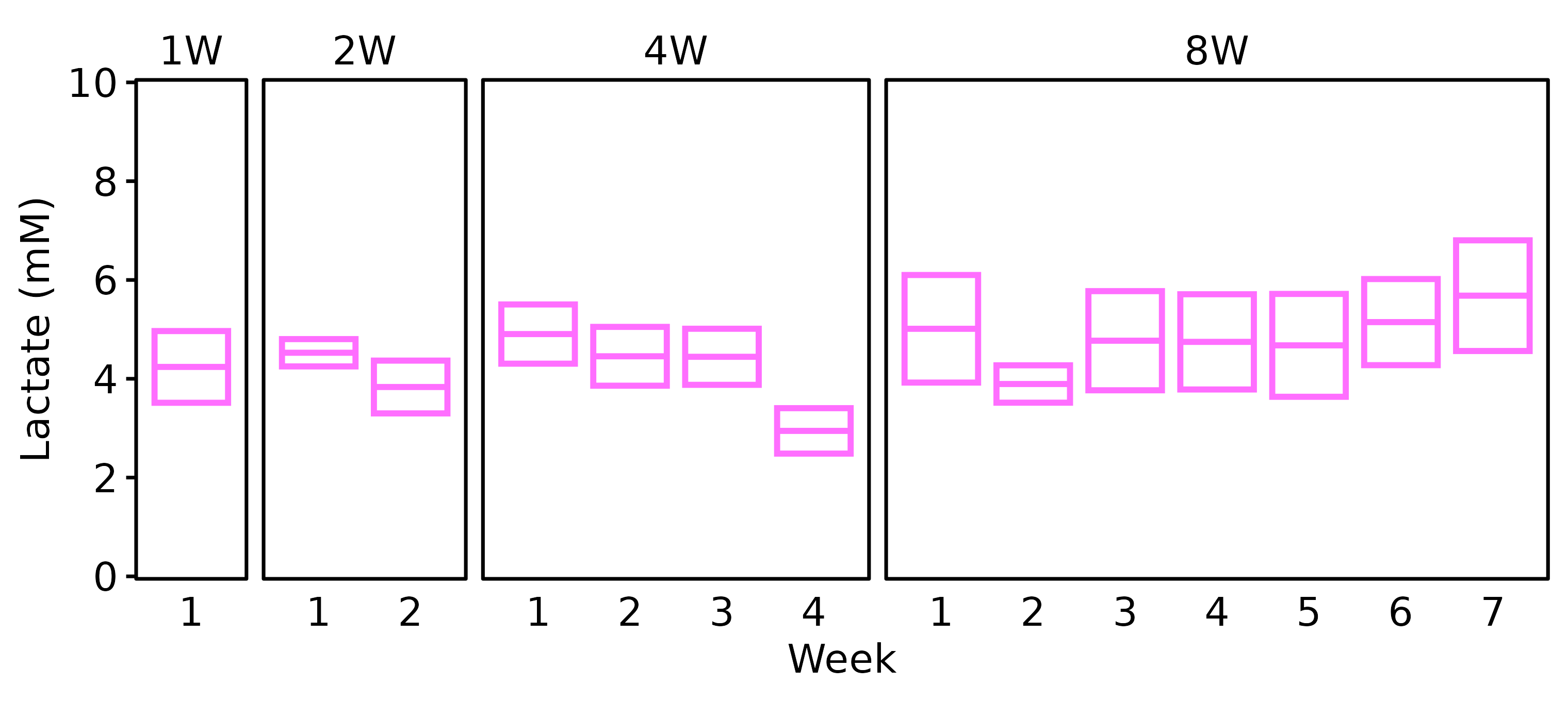
6M Male
# 6M Male
lact_df %>%
filter(age == "6M", sex == "Male") %>%
plot_lactate() +
scale_y_continuous(name = "Lactate (mM)",
expand = expansion(mult = 5e-3),
breaks = seq(0, 10, 2)) +
coord_cartesian(ylim = c(0, 10), clip = "off")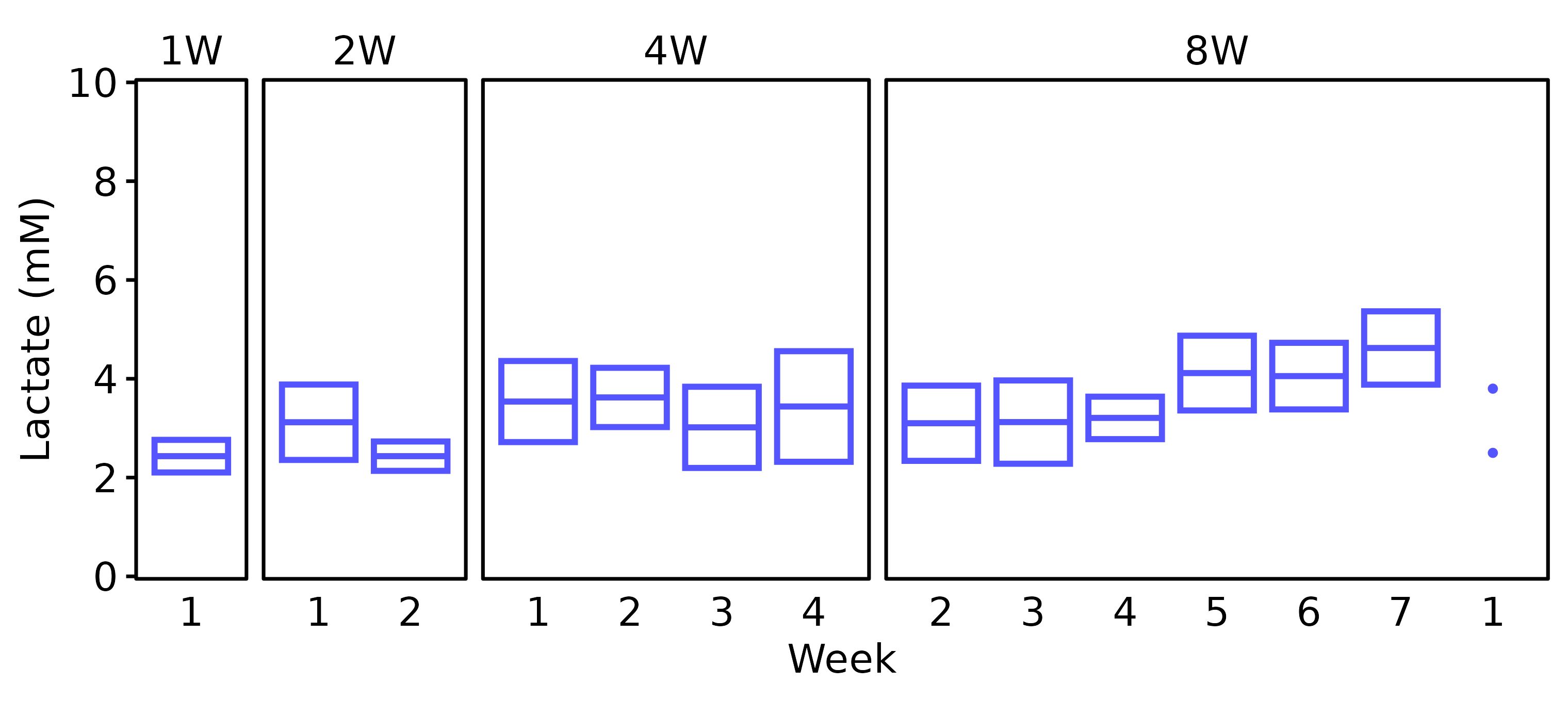
18M Female
# 18M Female
lact_df %>%
filter(age == "18M", sex == "Female") %>%
plot_lactate() +
scale_y_continuous(name = "Lactate (mM)",
expand = expansion(mult = 5e-3),
breaks = seq(0, 10, 2)) +
coord_cartesian(ylim = c(0, 10), clip = "off")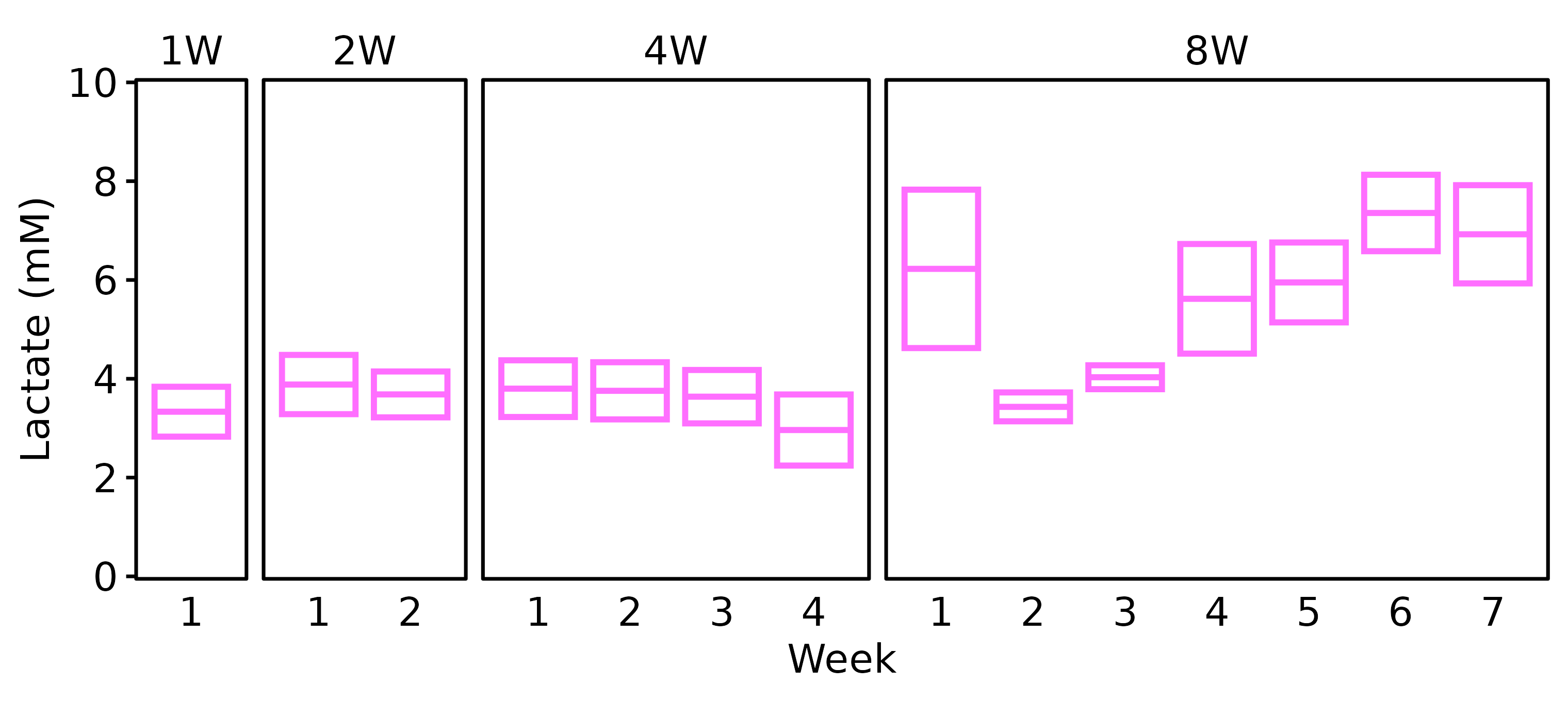
18M Male
# 18M Male
lact_df %>%
filter(age == "18M", sex == "Male") %>%
plot_lactate() +
scale_y_continuous(name = "Lactate (mM)",
expand = expansion(mult = 5e-3),
breaks = seq(0, 10, 2)) +
coord_cartesian(ylim = c(0, 10), clip = "off")