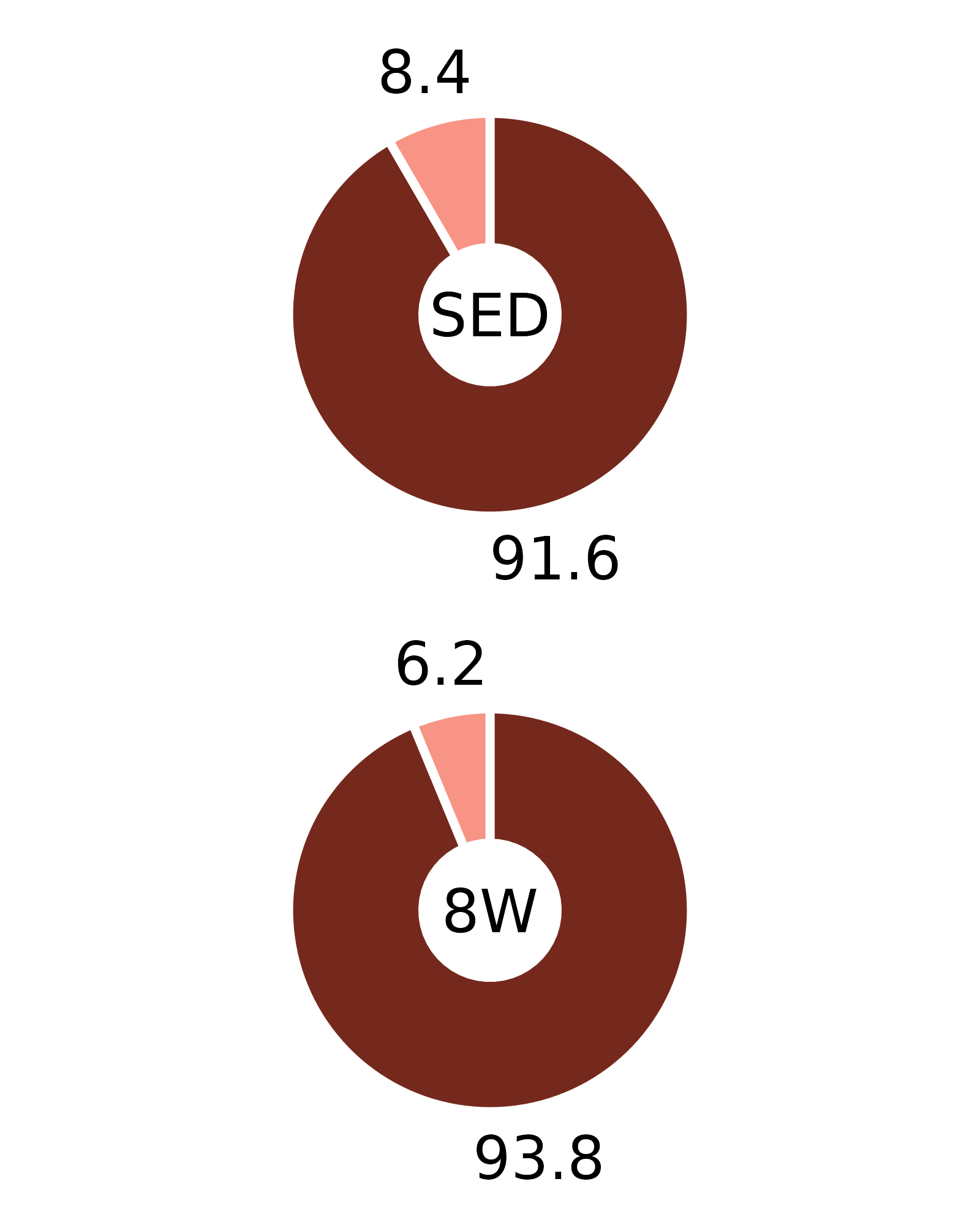
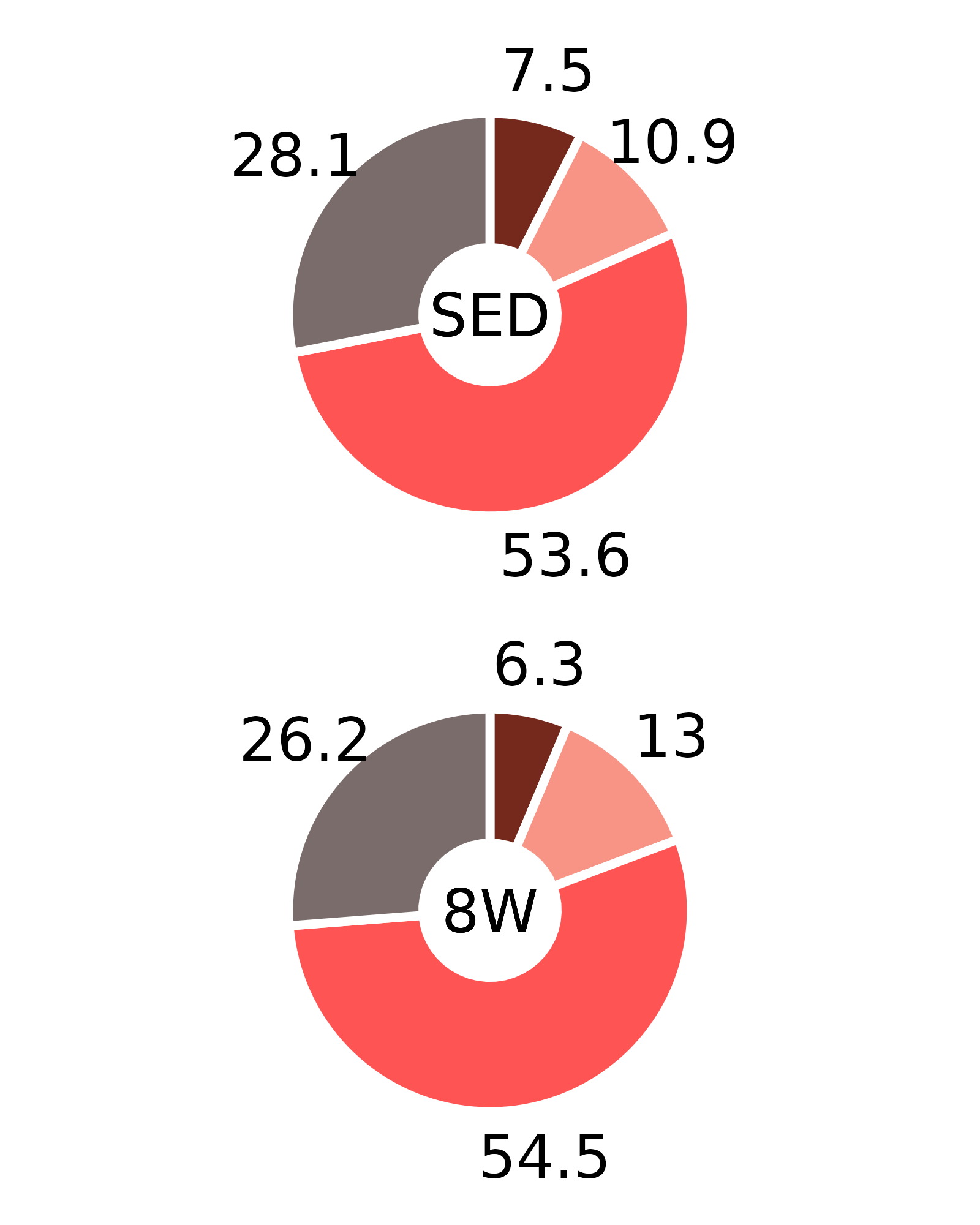
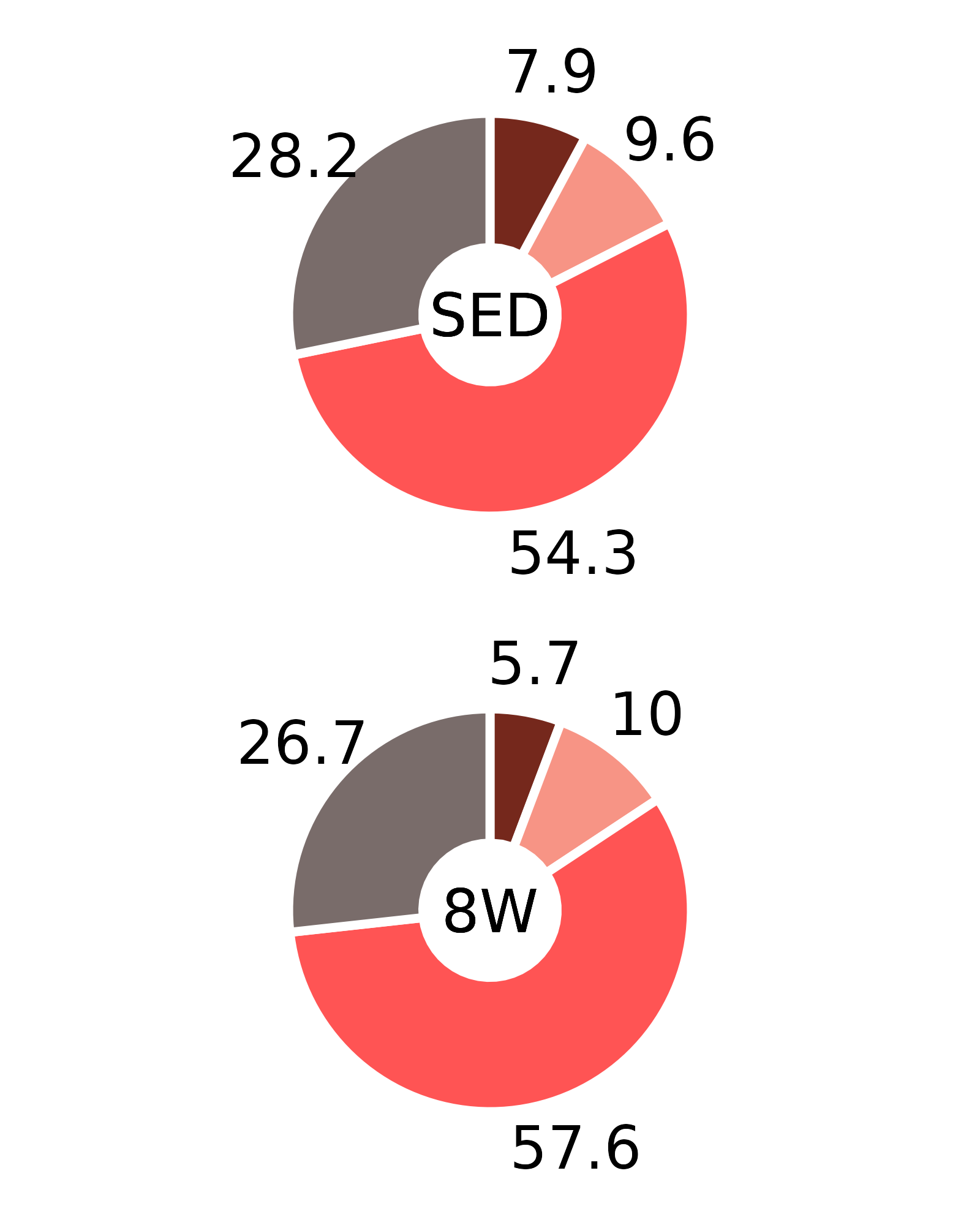
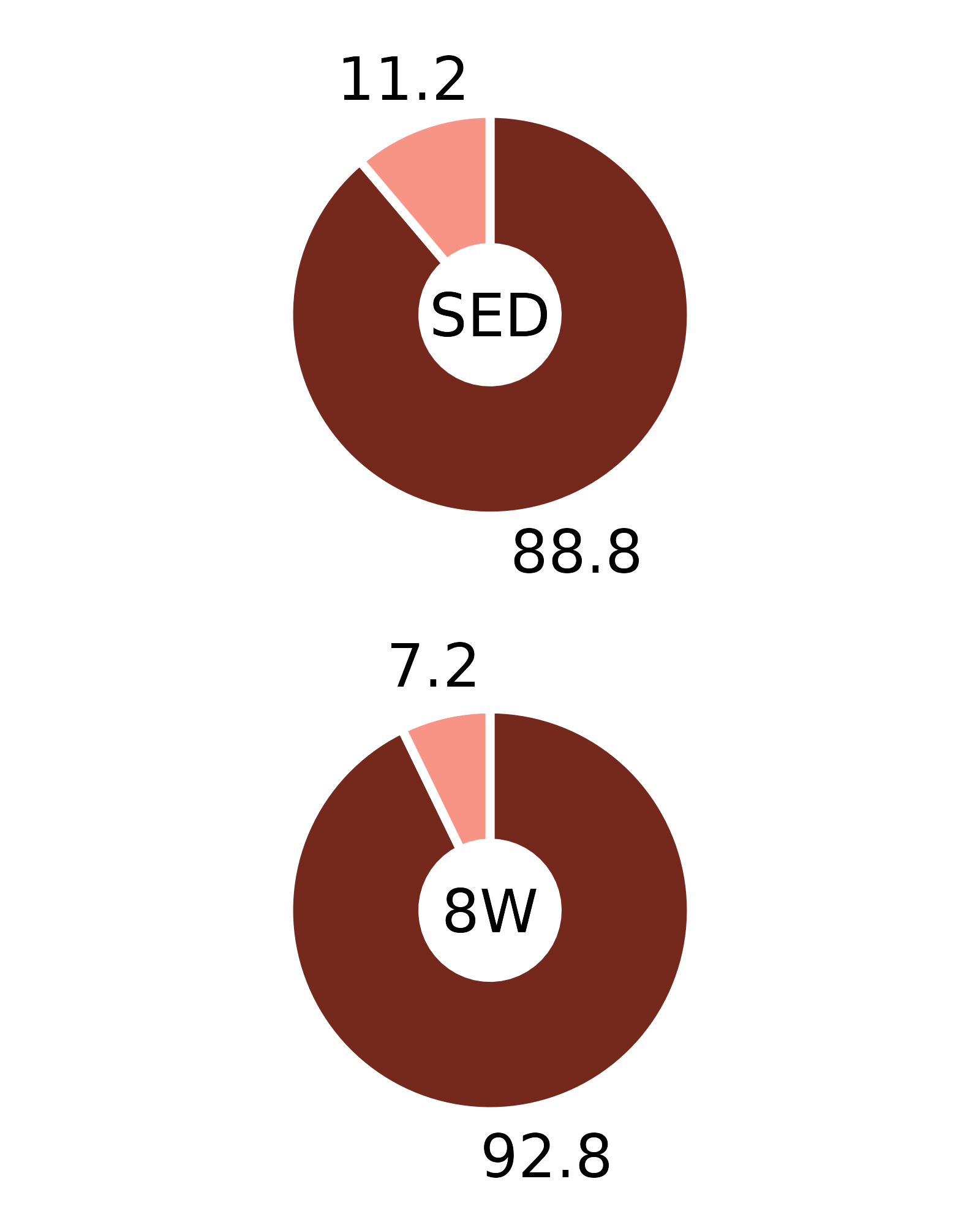
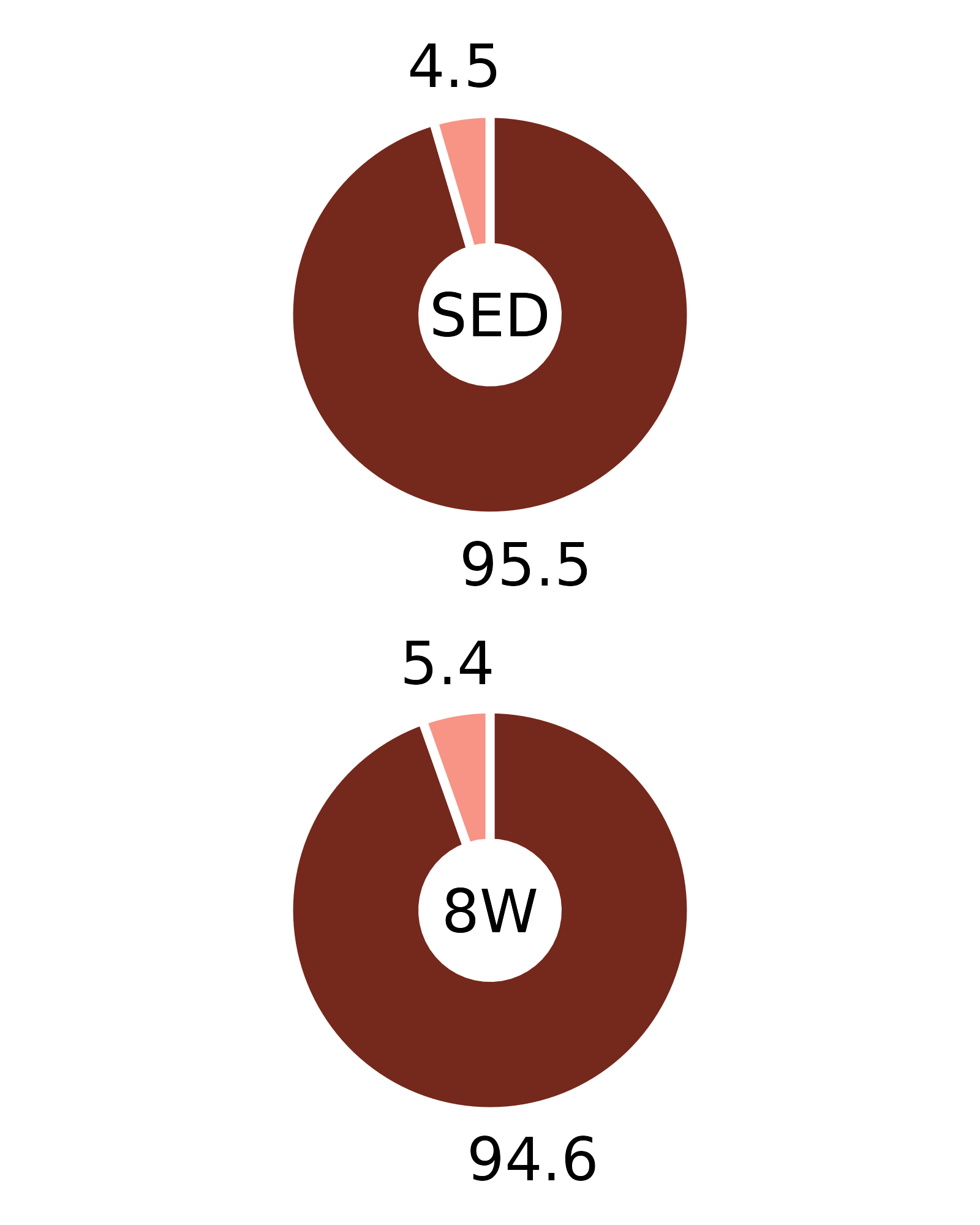
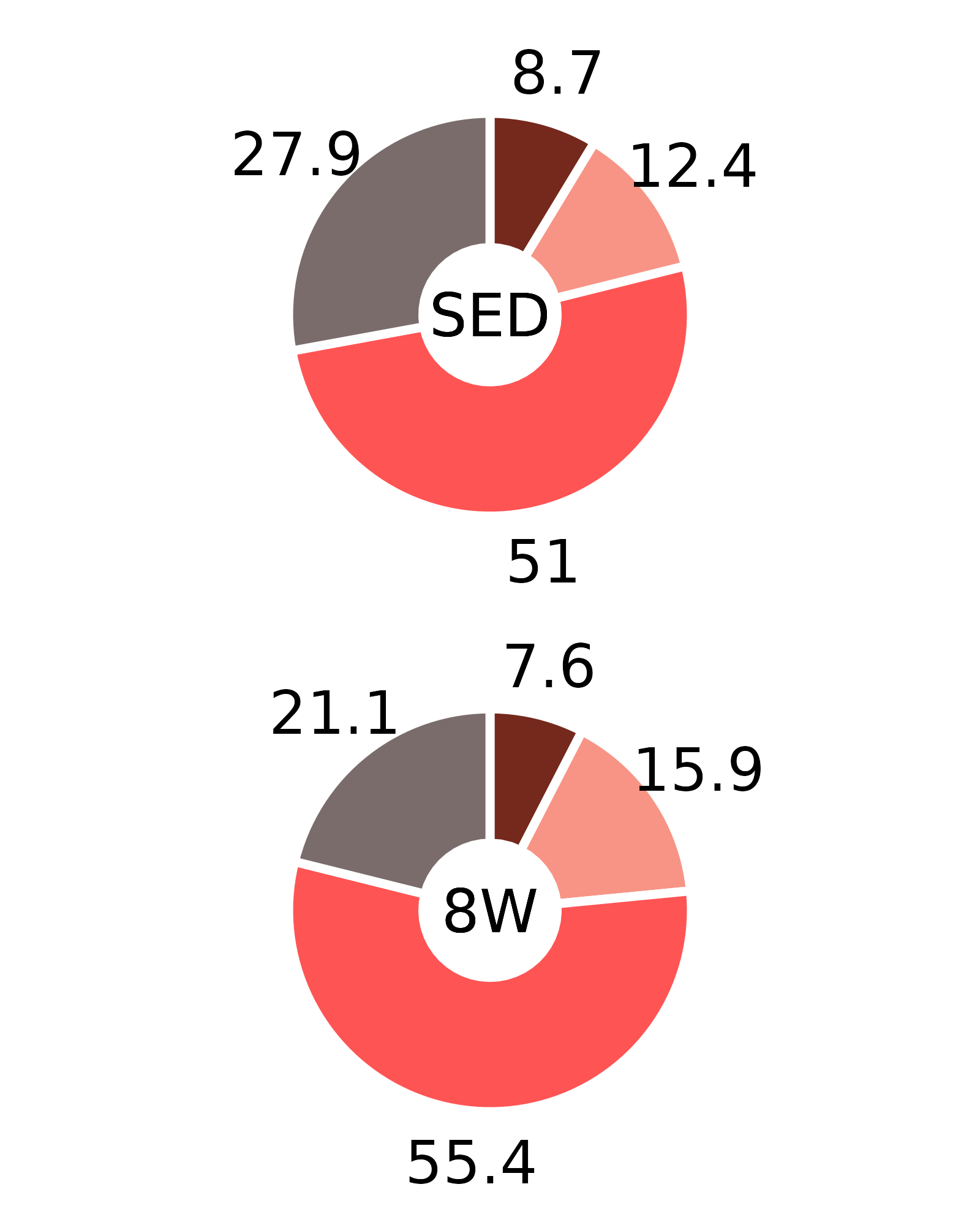
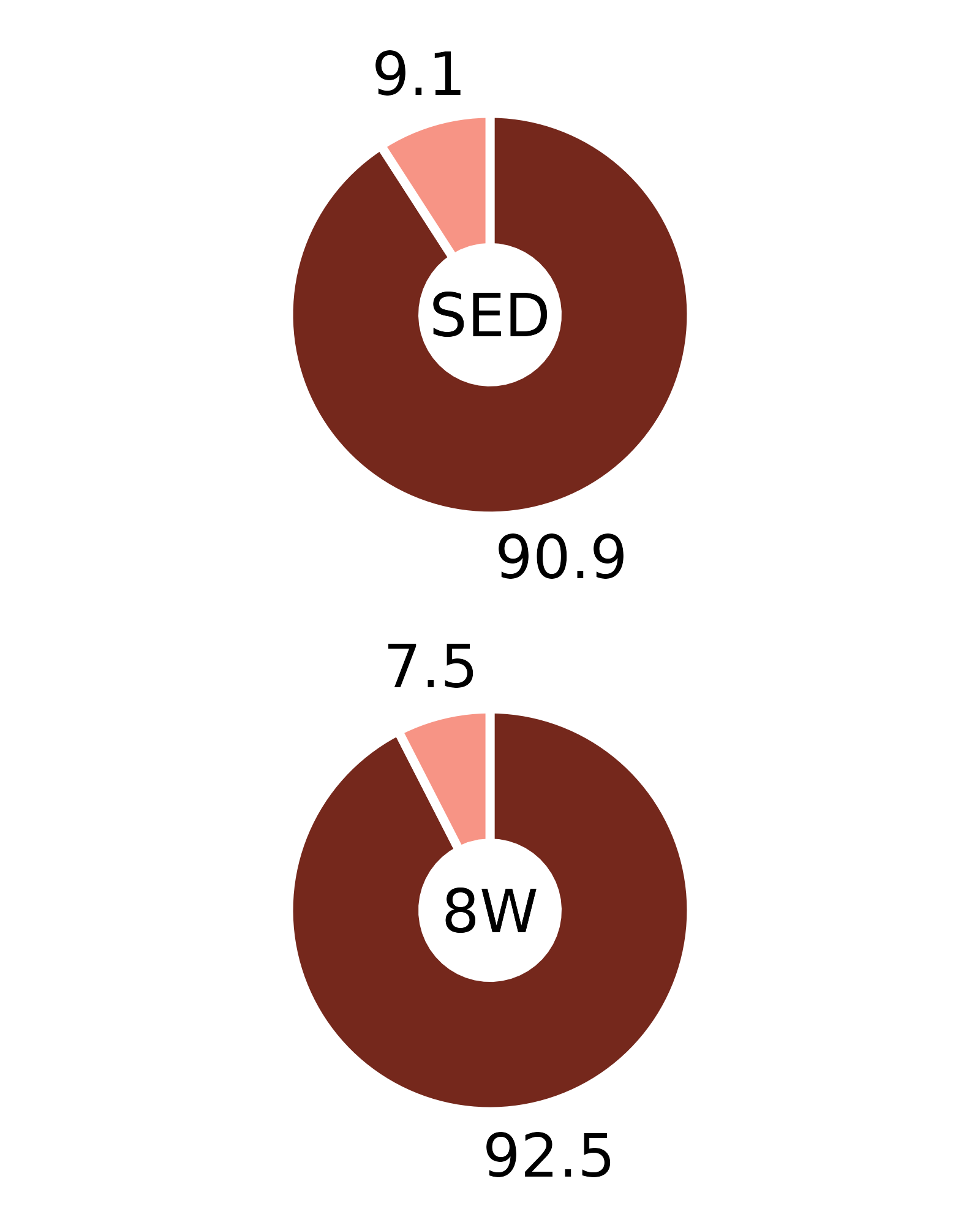
Plots of muscle fiber type percentages
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/fiber_count_plots.Rmd
fiber_count_plots.Rmd
# Required packages
library(MotrpacRatTrainingPhysiologyData)
library(dplyr)
library(ggplot2)
library(tidyr)
library(purrr)Overview
This article generates plots of the mean fiber type percentages. These values are calculated by summing the fiber counts across samples within each age, sex, group, and fiber type combination and dividing by the total fiber count and then multiplying by 100%. The percentage symbols (%) are not displayed on the plots to save space. Since there is no need to specify different y-axes, we can create and save all 16 plots with a few lines of code. Please refer to the “Statistical analysis of fiber counts by muscle and fiber type” vignette for details of the statistical analyses (not shown on the plots due to the complexity of the comparisons).
# Create all plots
foo <- FIBER_TYPES %>%
nest(.by = c(age, sex, muscle)) %>%
mutate(plots = map(.x = data, .f = fiber_donut_chart),
file_name = file.path("..", "..", "plots",
sprintf("fiber_count_%s_%s_%s.pdf",
muscle, age, tolower(sex))))
# Save all plots
map2(.x = foo$file_name, .y = foo$plots,
.f = ~ ggsave(filename = .x, plot = .y,
height = 2, width = 1.3,
family = "ArialMT"))
# Add legend back to plots for display and to save legend separately
foo <- foo %>%
mutate(plots = map(.x = plots, .f = ~ .x + theme(legend.position = "right")))
# Save legend
lgd <- get_legend(p = foo$plots[[1]]) %>%
as_ggplot()
ggsave(filename = file.path("..", "..", "plots", "fiber_count_legend.pdf"),
plot = lgd, height = 0.7, width = 0.4, family = "ArialMT")Session Info
sessionInfo()
#> R version 4.4.0 (2024-04-24)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] purrr_1.0.2
#> [2] tidyr_1.3.1
#> [3] ggplot2_3.5.1
#> [4] dplyr_1.1.4
#> [5] MotrpacRatTrainingPhysiologyData_2.0.0
#>
#> loaded via a namespace (and not attached):
#> [1] sass_0.4.9 utf8_1.2.4 generics_0.1.3 rstatix_0.7.2
#> [5] digest_0.6.35 magrittr_2.0.3 evaluate_0.23 grid_4.4.0
#> [9] fastmap_1.1.1 jsonlite_1.8.8 backports_1.4.1 fansi_1.0.6
#> [13] scales_1.3.0 textshaping_0.3.7 jquerylib_0.1.4 abind_1.4-5
#> [17] cli_3.6.2 rlang_1.1.3 munsell_0.5.1 withr_3.0.0
#> [21] cachem_1.0.8 yaml_2.3.8 ggbeeswarm_0.7.2 tools_4.4.0
#> [25] memoise_2.0.1 ggsignif_0.6.4 colorspace_2.1-0 ggpubr_0.6.0
#> [29] broom_1.0.5 vctrs_0.6.5 R6_2.5.1 lifecycle_1.0.4
#> [33] fs_1.6.4 car_3.1-2 htmlwidgets_1.6.4 vipor_0.4.7
#> [37] ragg_1.3.0 pkgconfig_2.0.3 beeswarm_0.4.0 desc_1.4.3
#> [41] pkgdown_2.0.9 pillar_1.9.0 bslib_0.7.0 gtable_0.3.5
#> [45] glue_1.7.0 systemfonts_1.0.6 highr_0.10 xfun_0.43
#> [49] tibble_3.2.1 tidyselect_1.2.1 knitr_1.46 farver_2.1.1
#> [53] htmltools_0.5.8.1 labeling_0.4.3 rmarkdown_2.26 carData_3.0-5
#> [57] compiler_4.4.0