Plots of muscle-specific measures
Tyler Sagendorf
01 May, 2024
Source:vignettes/articles/muscle_plots.Rmd
muscle_plots.Rmd
# Required packages
library(MotrpacRatTrainingPhysiologyData)
library(dplyr)
library(tibble)
library(tidyr)
library(purrr)
library(ggplot2)
library(latex2exp)
library(grid)Overview
This article generates plots for all muscle-specific measures:
glycogen, citrate synthase, terminal muscle mass, cross-sectional area,
and capillary contacts. Brackets indicate a statistically significant
difference between the means of the SED group and a trained group (1W,
2W, 4W, 8W). A significant difference is defined as 1) a Dunnett’s test
p-value < 0.05 for measures of glycogen, citrate synthase, and
terminal muscle mass or 2) a Holm-adjusted p-value < 0.05 for
measures of cross-sectional area and capillary contacts. Please refer to
the “Statistical analyses of muscle-specific measures” vignette for code
to generate MUSCLES_STATS.
Data Preparation:
We apply the summary method to each emmGrid
object and rename columns to work with plot_baseline. This
list of objects is then converted to a single data.frame
with the response column specifying which measurement is
being tested.
# Reformat confidence interval data
conf_df <- map(MUSCLES_EMM, function(emm_i) {
terms_i <- attr(terms(emm_i@model.info), which = "term.labels")
out <- summary(emm_i) %>%
as.data.frame() %>%
# Rename columns for use in plot_baseline
rename(any_of(c(lower.CL = "asymp.LCL",
upper.CL = "asymp.UCL",
response_mean = "response",
response_mean = "rate"))) %>%
mutate(group = factor(group, levels = c("SED", paste0(2 ^ (0:3), "W"))))
return(out)
}) %>%
enframe(name = "response") %>%
unnest(value)Glycogen
Muscle glycogen concentration (ng glycogen/\(\mu\)L supernatant/mg tissue).
LG
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "LG"),
sex = "Female", age = "6M", y_position = 4.7) +
scale_y_continuous(name = TeX("LG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 12, 2),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 12), clip = "off")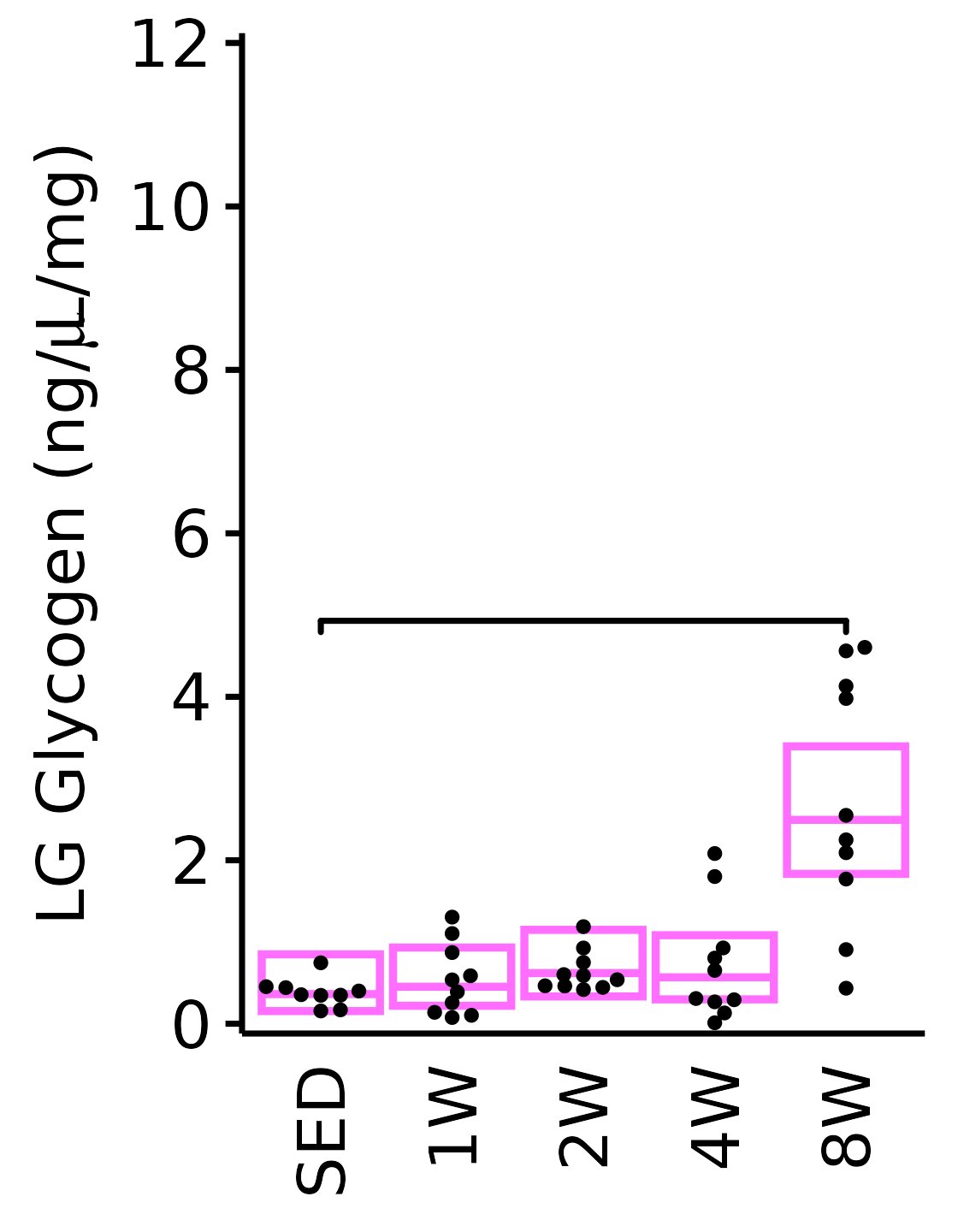
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "LG"),
sex = "Male", age = "6M", y_position = 9.5) +
scale_y_continuous(name = TeX("LG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 16, 4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 16), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))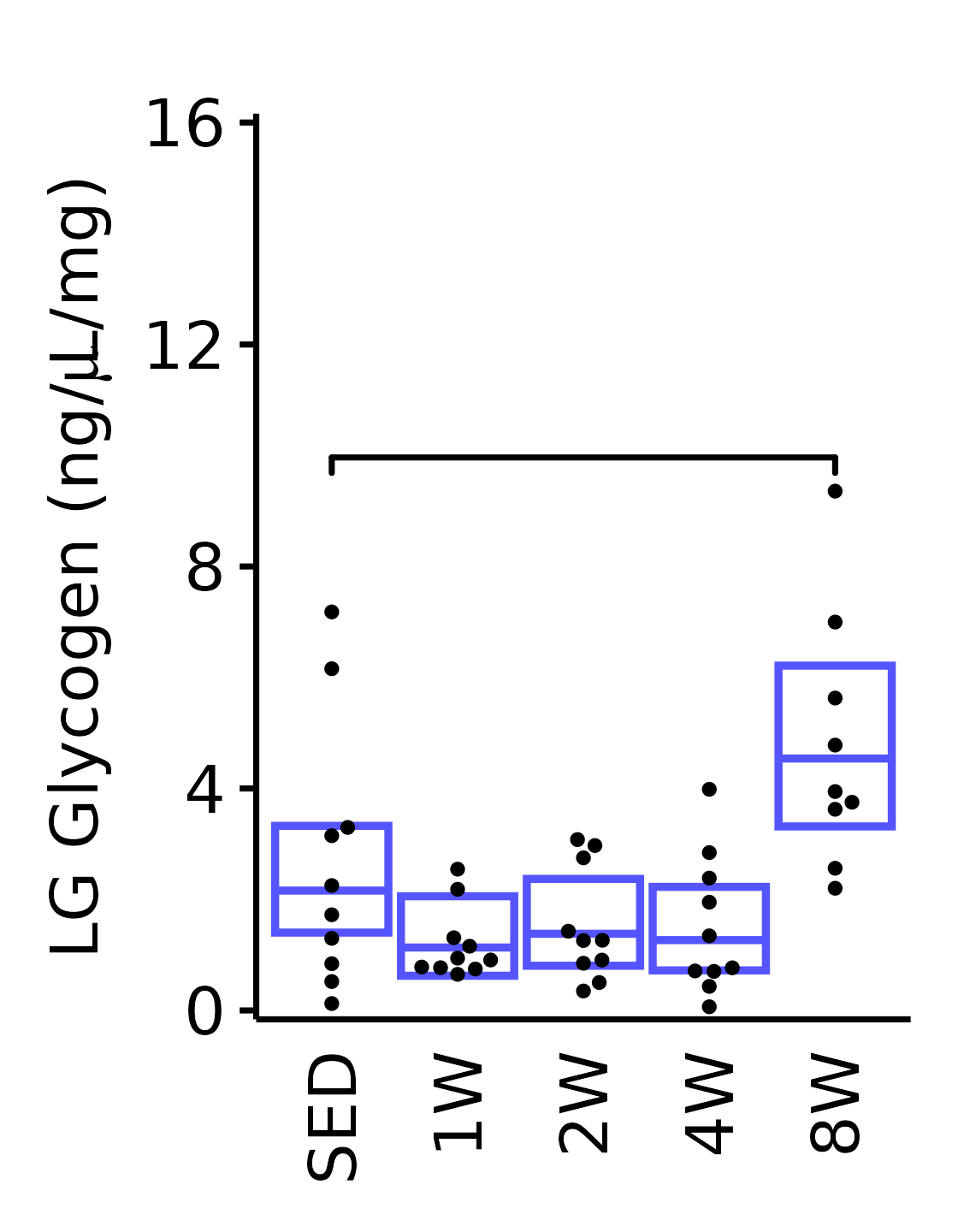
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "LG"),
sex = "Female", age = "18M") +
scale_y_continuous(name = TeX("LG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 12, 2),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 12), clip = "off")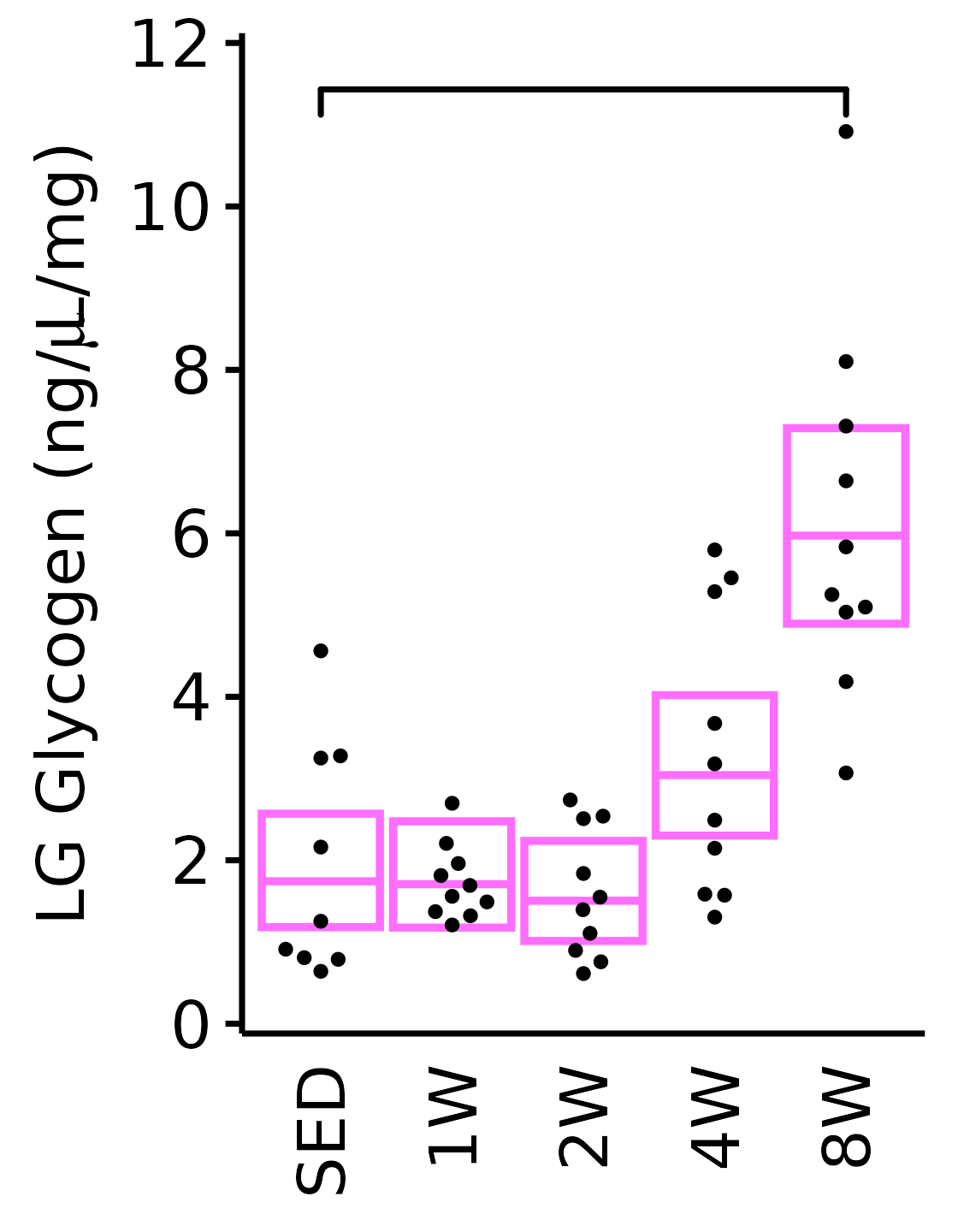
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "LG"),
sex = "Male", age = "18M", y_position = 16.7) +
scale_y_continuous(name = TeX("LG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 16, 4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 16), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))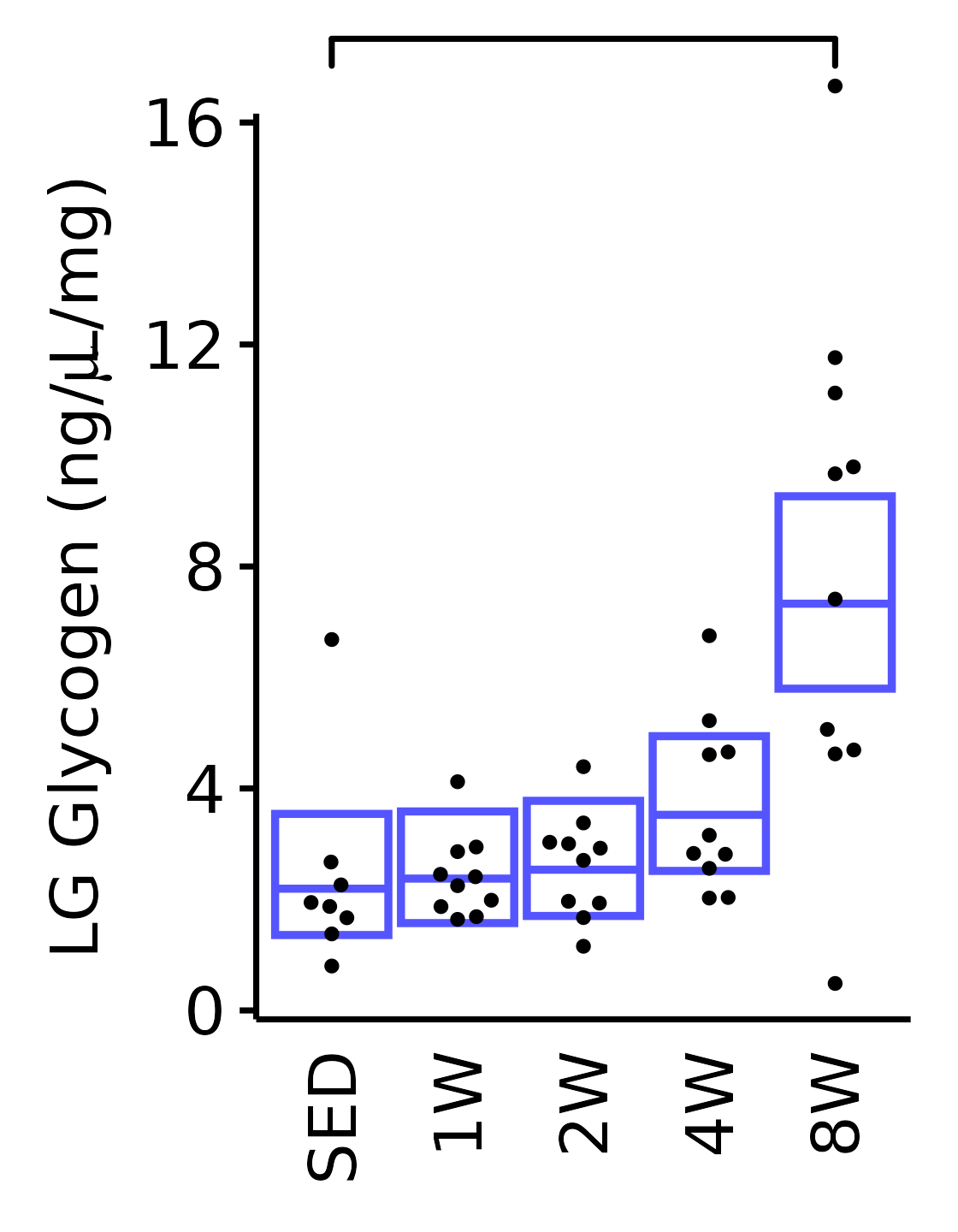
MG
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "MG"),
sex = "Female", age = "6M") +
scale_y_continuous(name = TeX("MG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 12, 4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 13), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))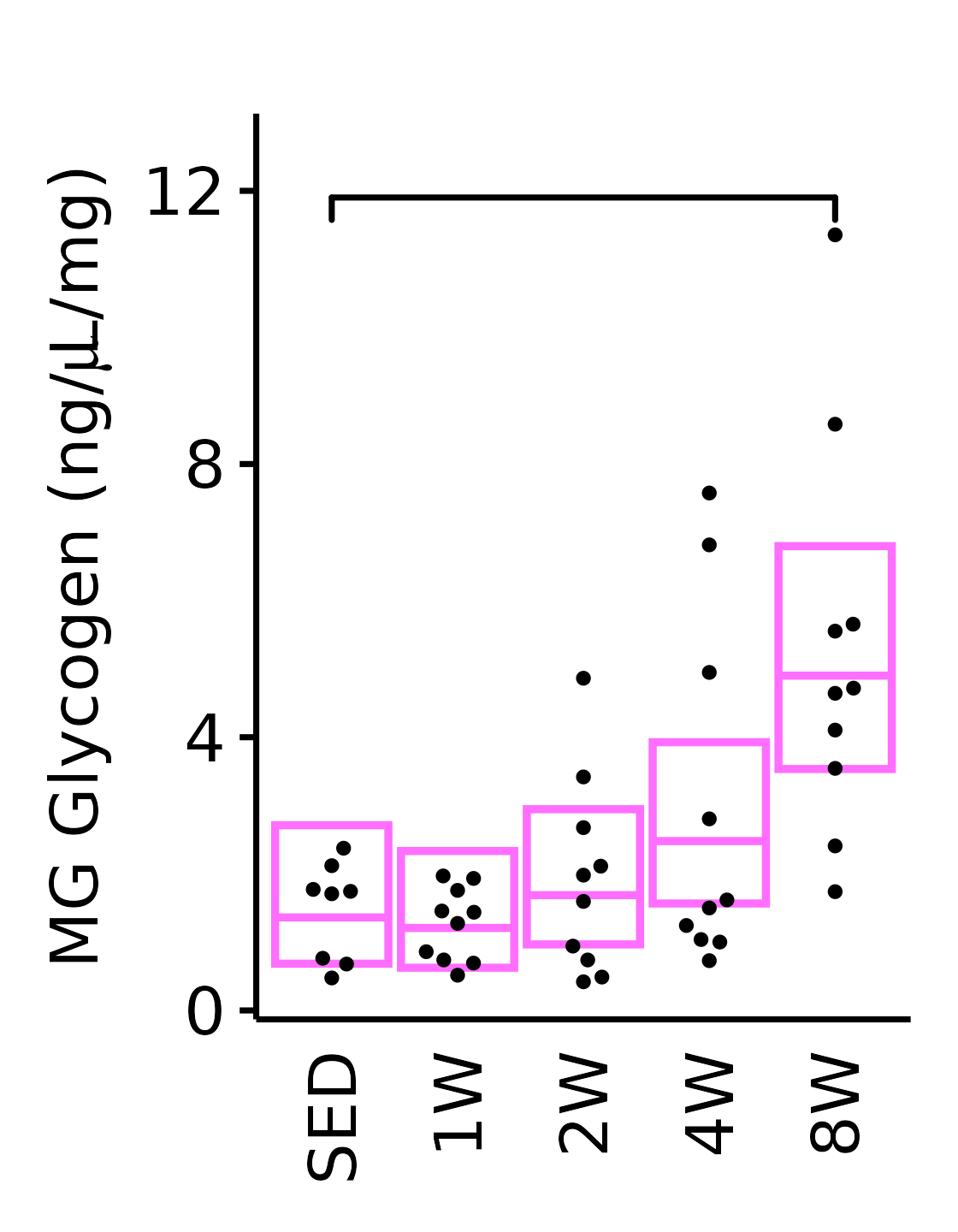
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "MG"),
sex = "Male", age = "6M") +
scale_y_continuous(name = TeX("MG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 20, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 20), clip = "off")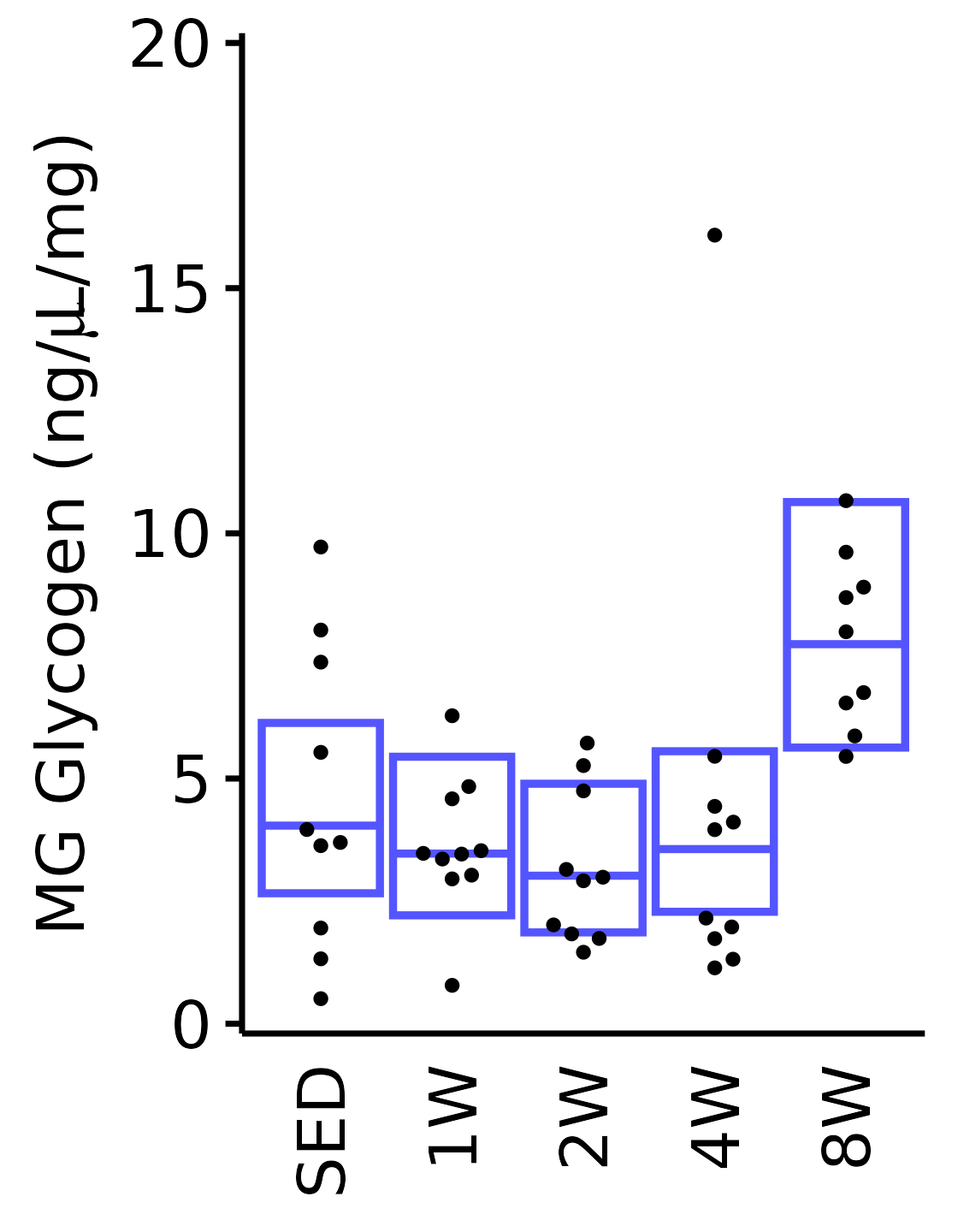
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "MG"),
sex = "Female", age = "18M", y_position = 12.4) +
scale_y_continuous(name = TeX("MG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 12, 4),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 13), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))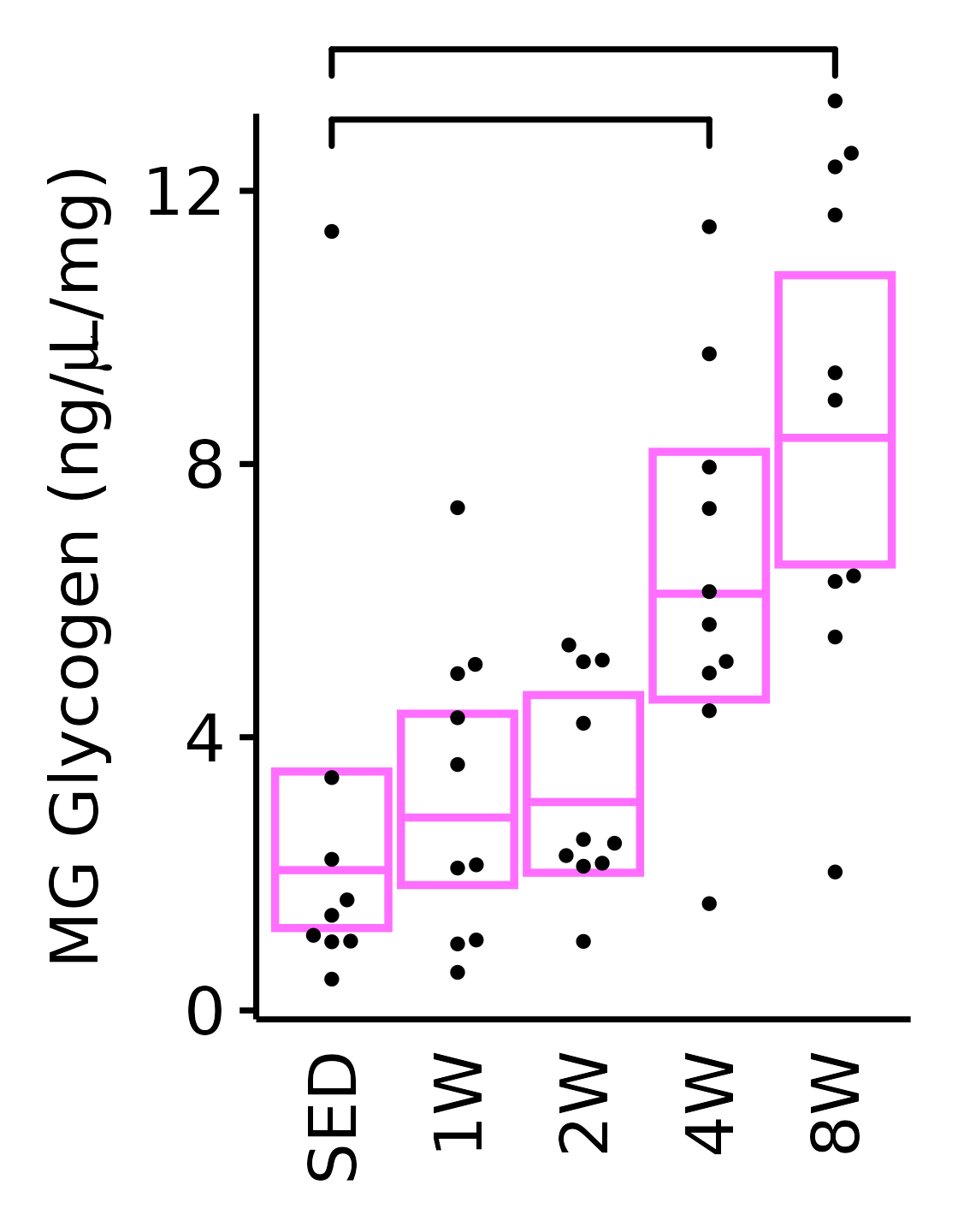
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "MG"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("MG Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 20, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 20), clip = "off")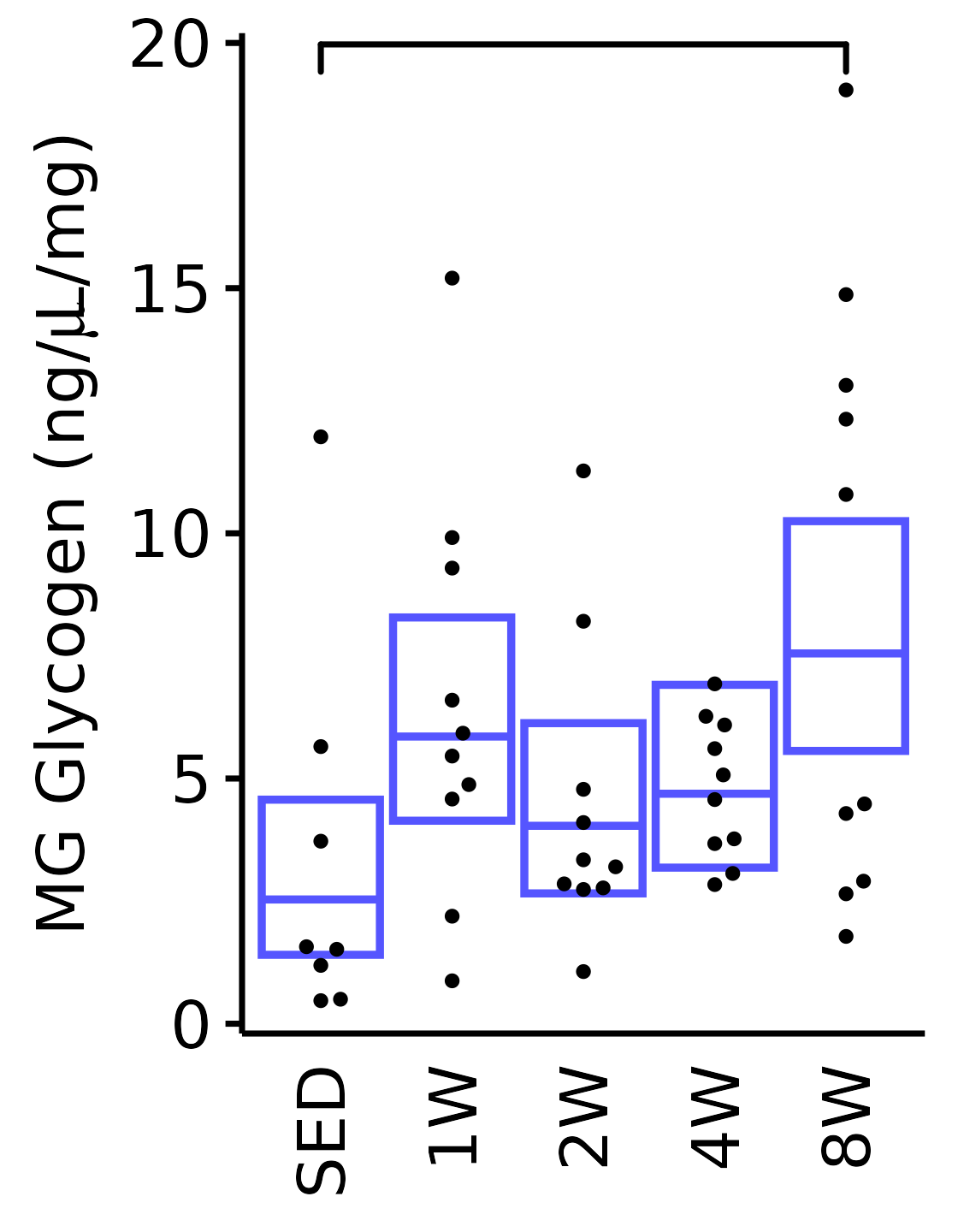
PL
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "PL"),
sex = "Female", age = "6M", y_position = 14) +
scale_y_continuous(name = TeX("PL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 30, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 30), clip = "off")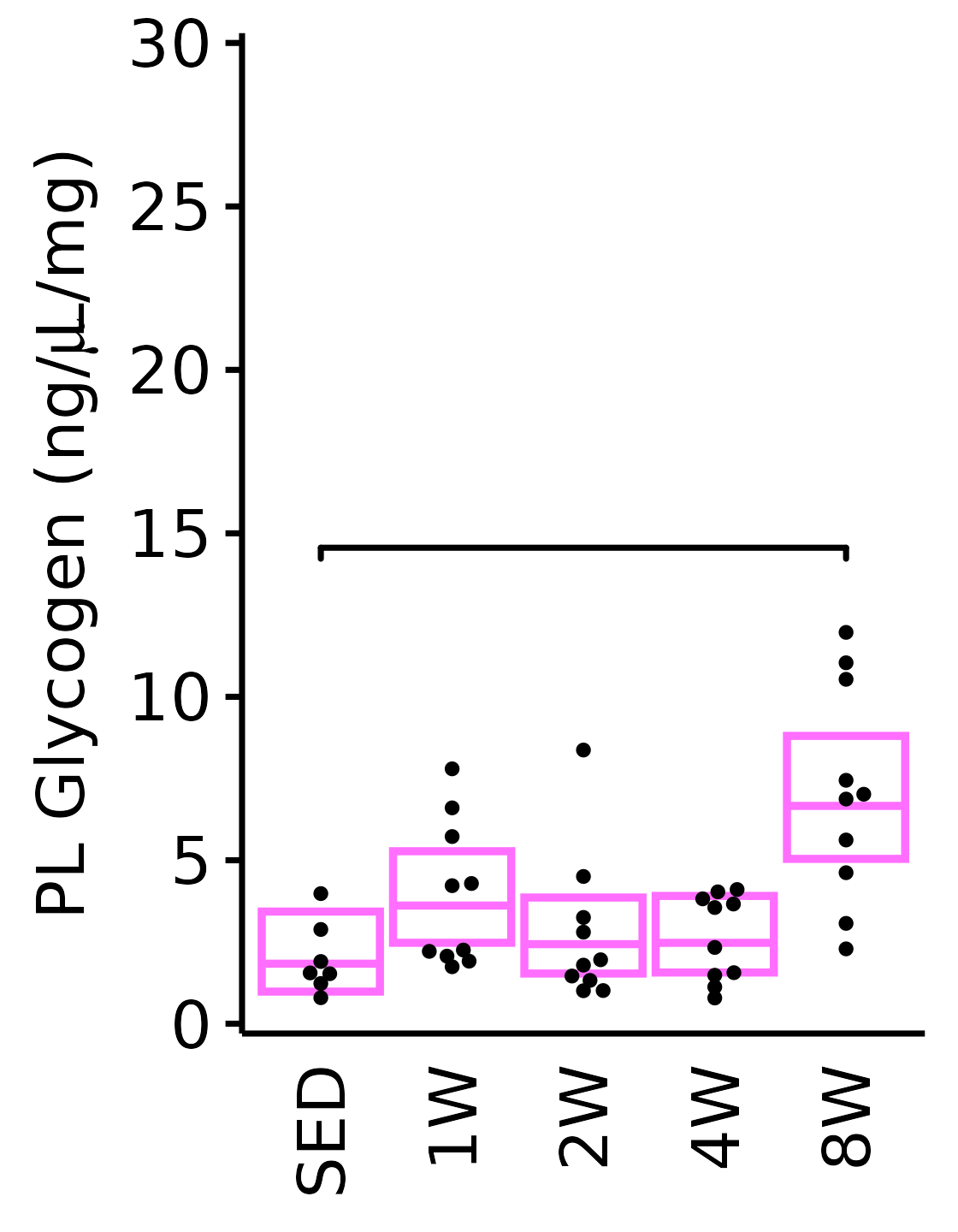
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "PL"),
sex = "Male", age = "6M", y_position = 13) +
scale_y_continuous(name = TeX("PL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 25, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 25), clip = "off")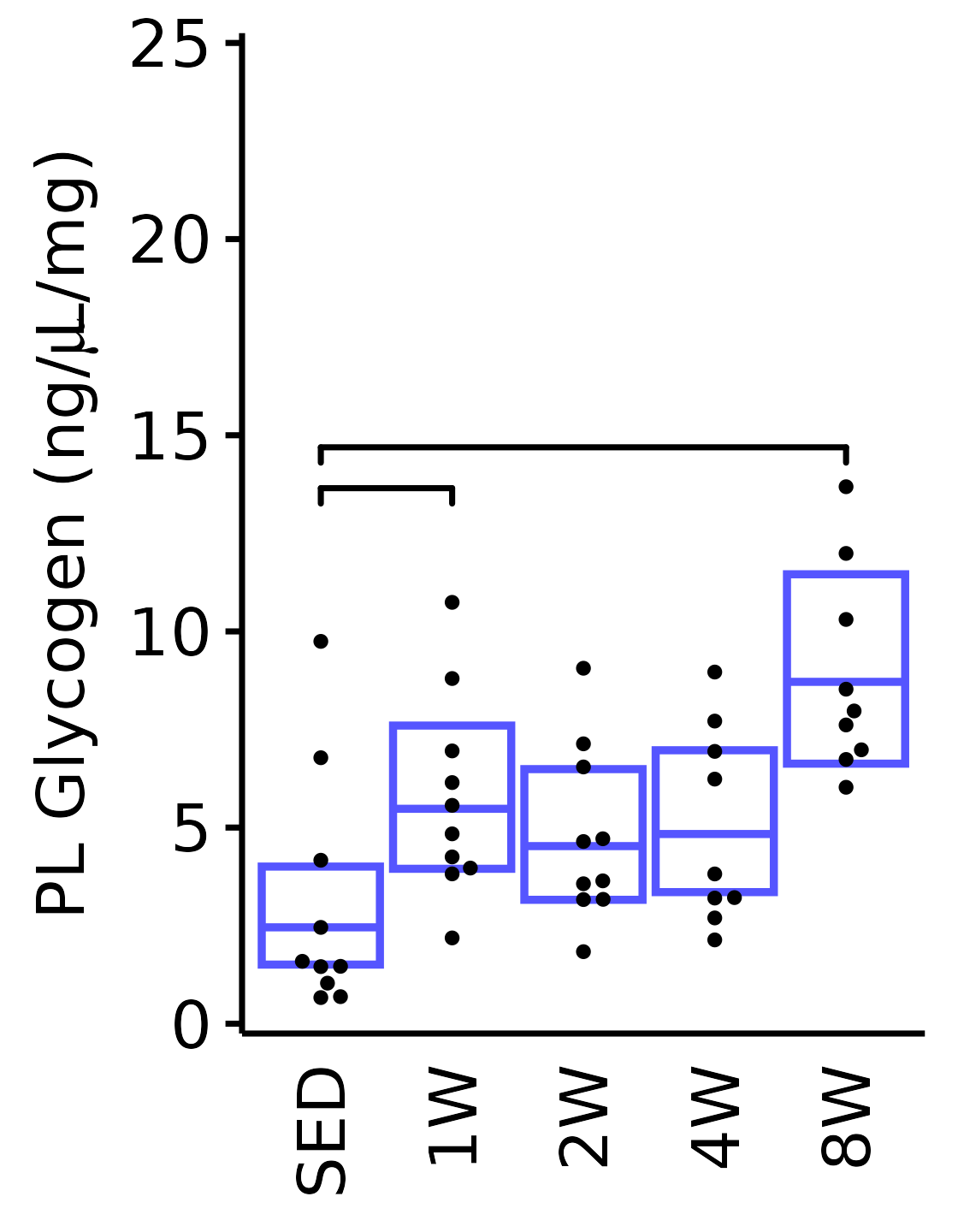
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "PL"),
sex = "Female", age = "18M") +
scale_y_continuous(name = TeX("PL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 30, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 30), clip = "off")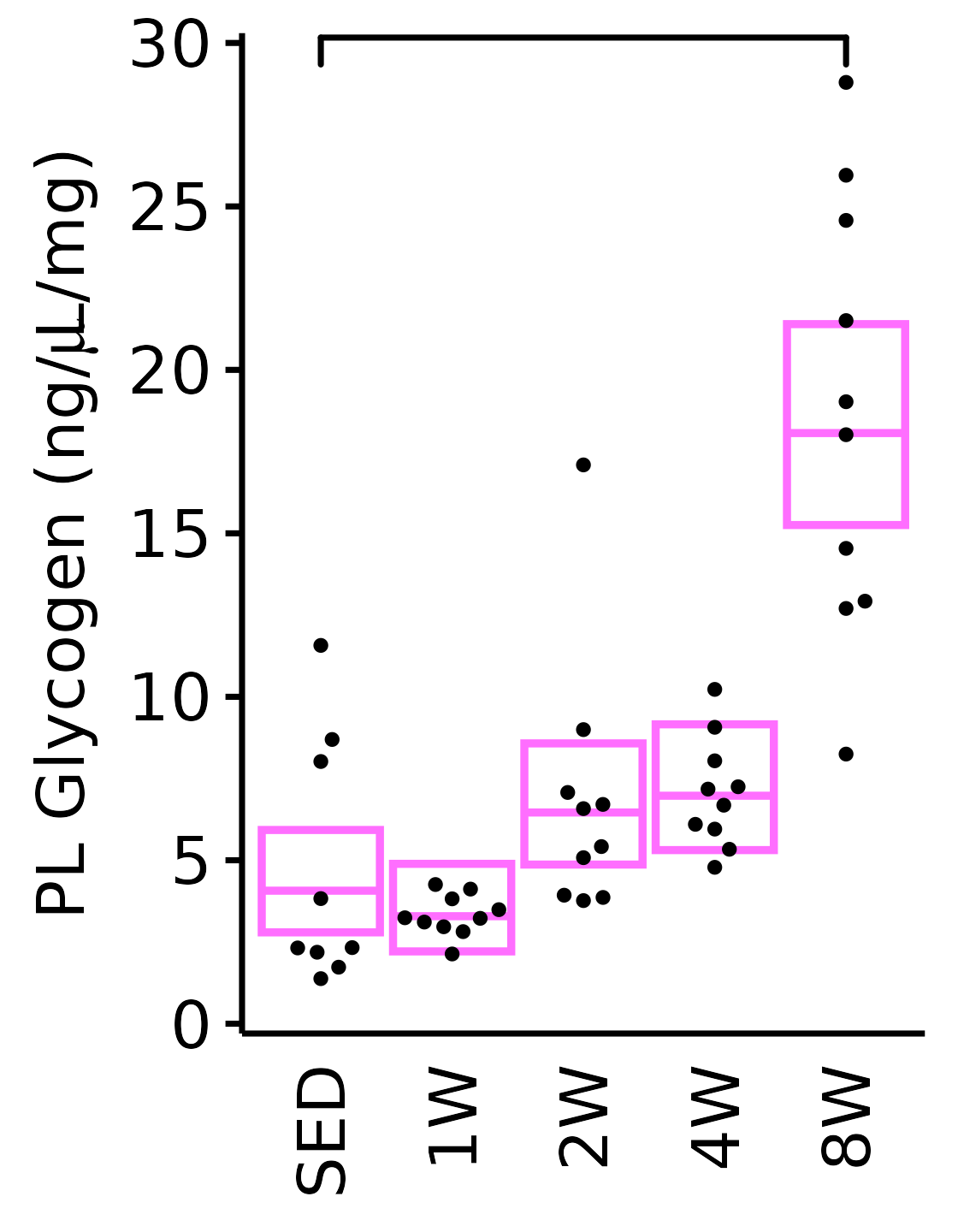
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "PL"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("PL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 25, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 25), clip = "off")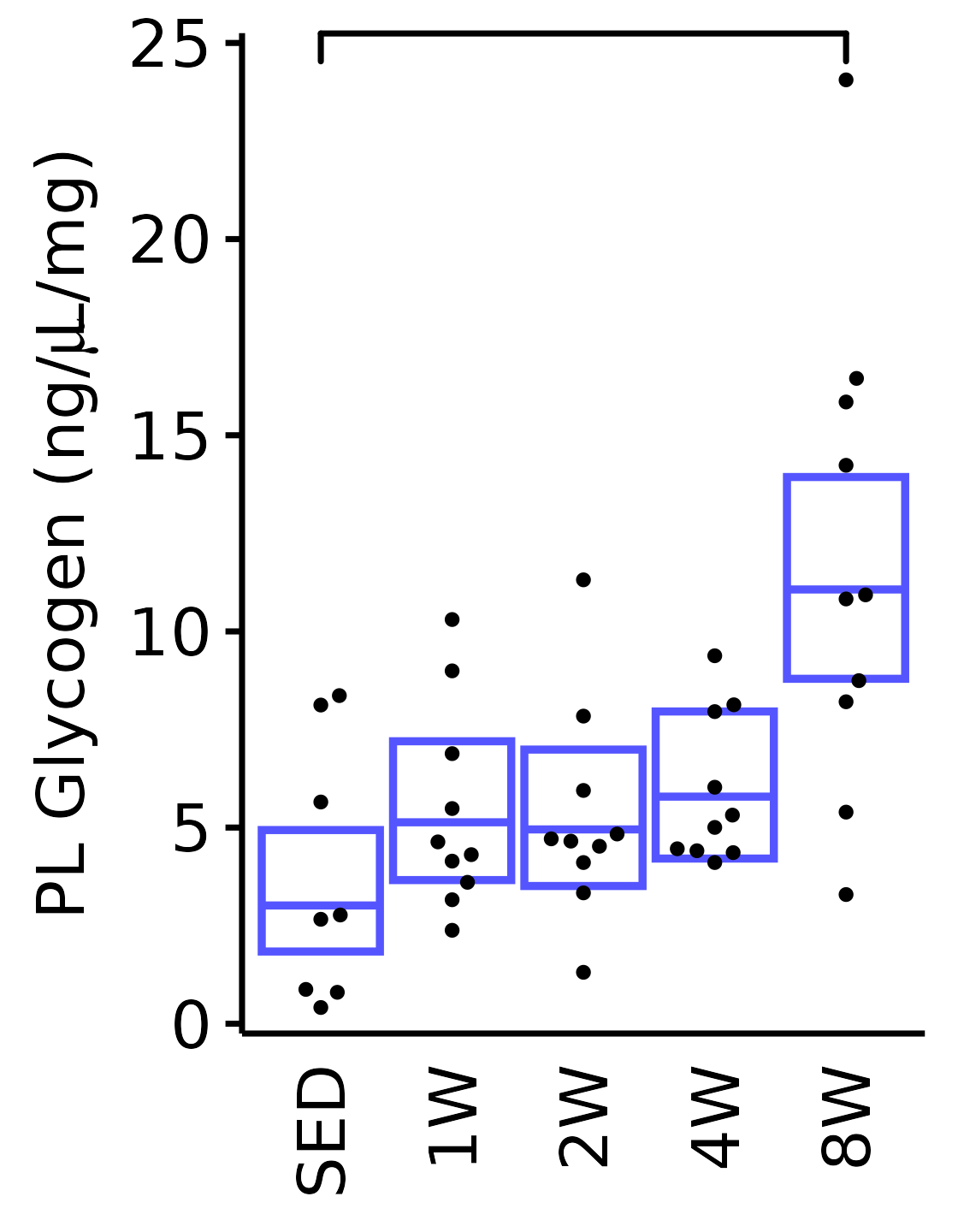
SOL
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "SOL"),
sex = "Female", age = "6M") +
scale_y_continuous(name = TeX("SOL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 60, 10),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 64), clip = "off")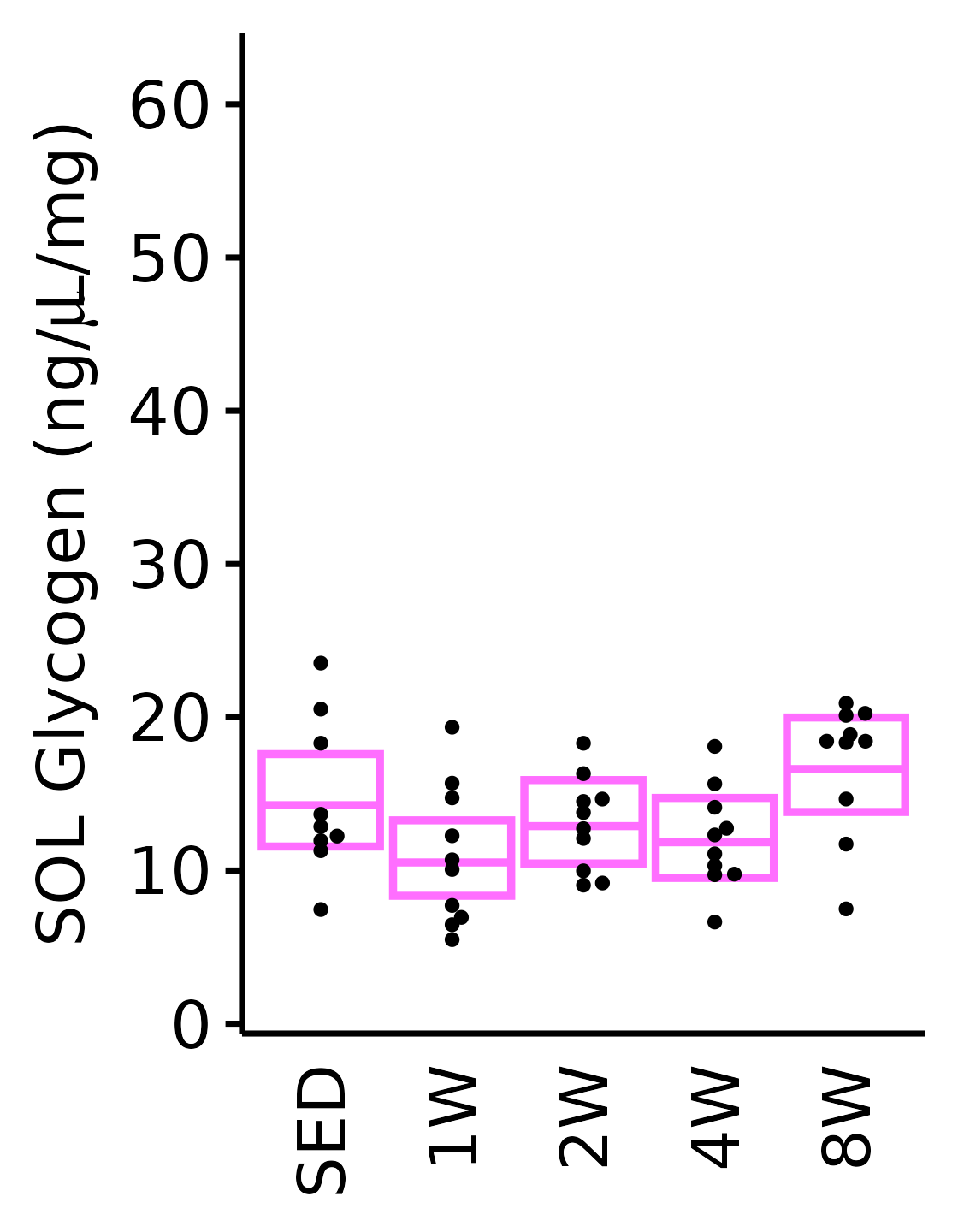
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "SOL"),
sex = "Male", age = "6M", y_position = 27) +
scale_y_continuous(name = TeX("SOL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 35, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 35), clip = "off")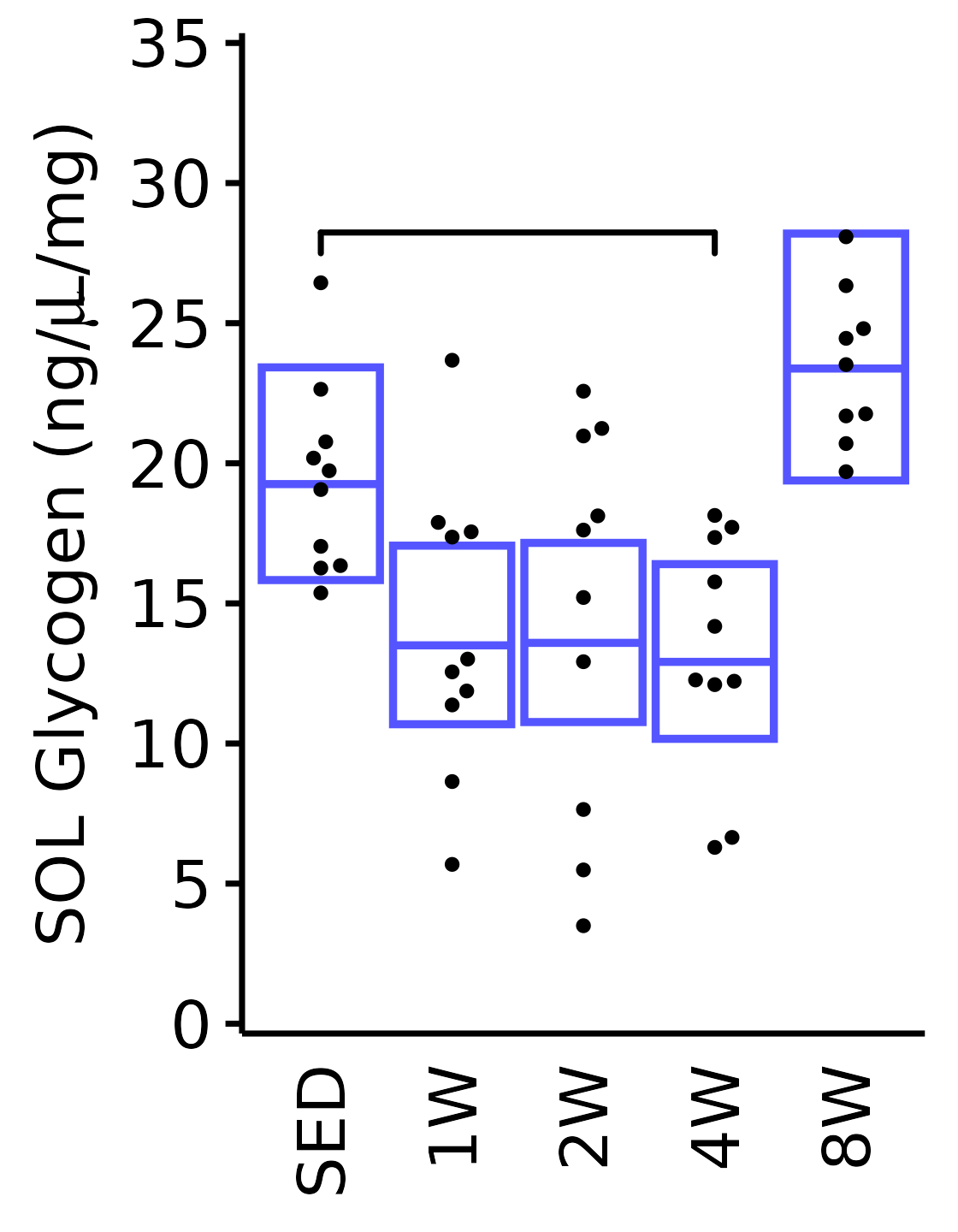
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "SOL"),
sex = "Female", age = "18M", y_position = 50) +
scale_y_continuous(name = TeX("SOL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 60, 10),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 64), clip = "off")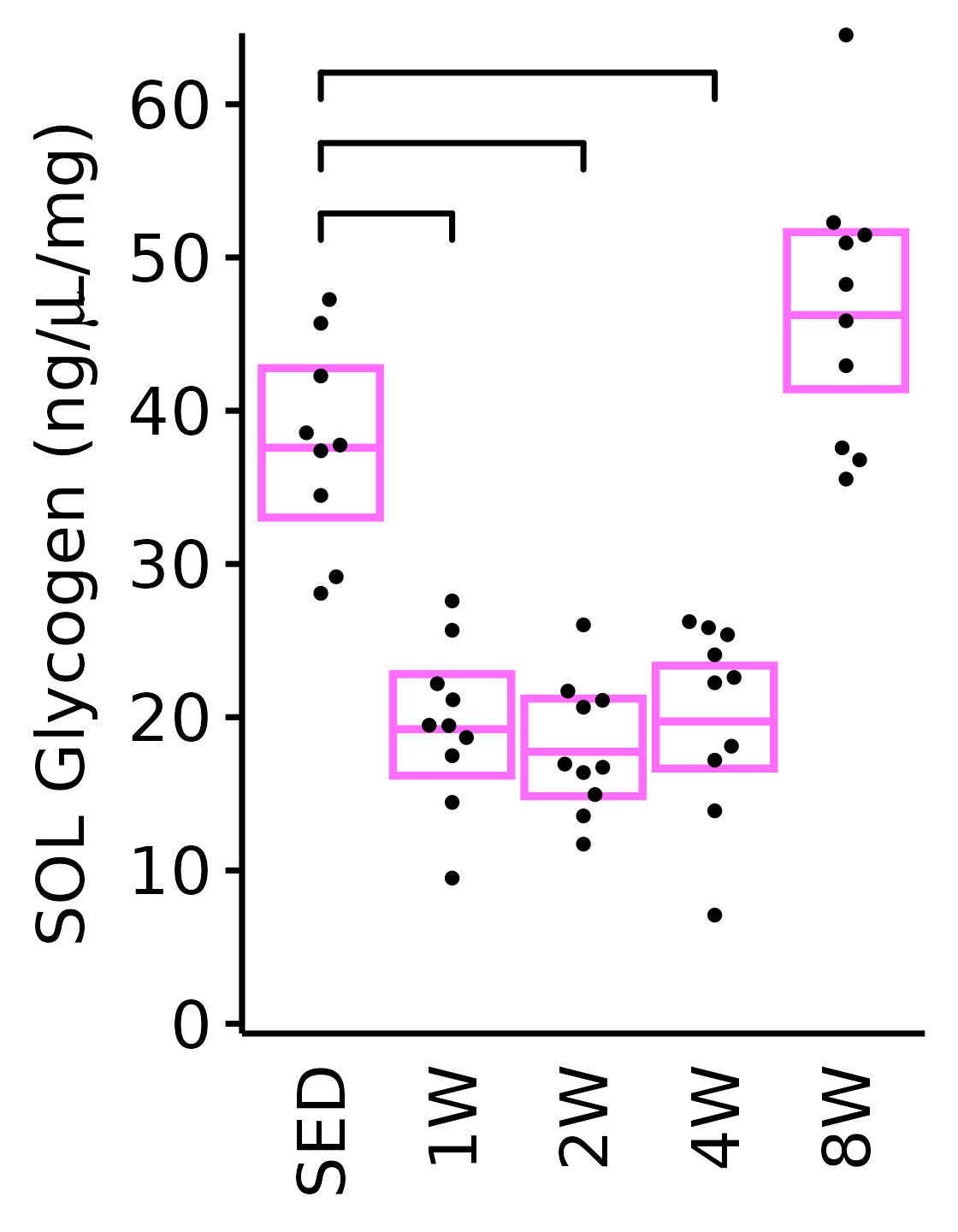
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "glycogen",
conf = filter(conf_df, response == "Glycogen",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Glycogen",
muscle == "SOL"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("SOL Glycogen (ng/$\\mu$L/mg)"),
breaks = seq(0, 35, 5),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 35), clip = "off")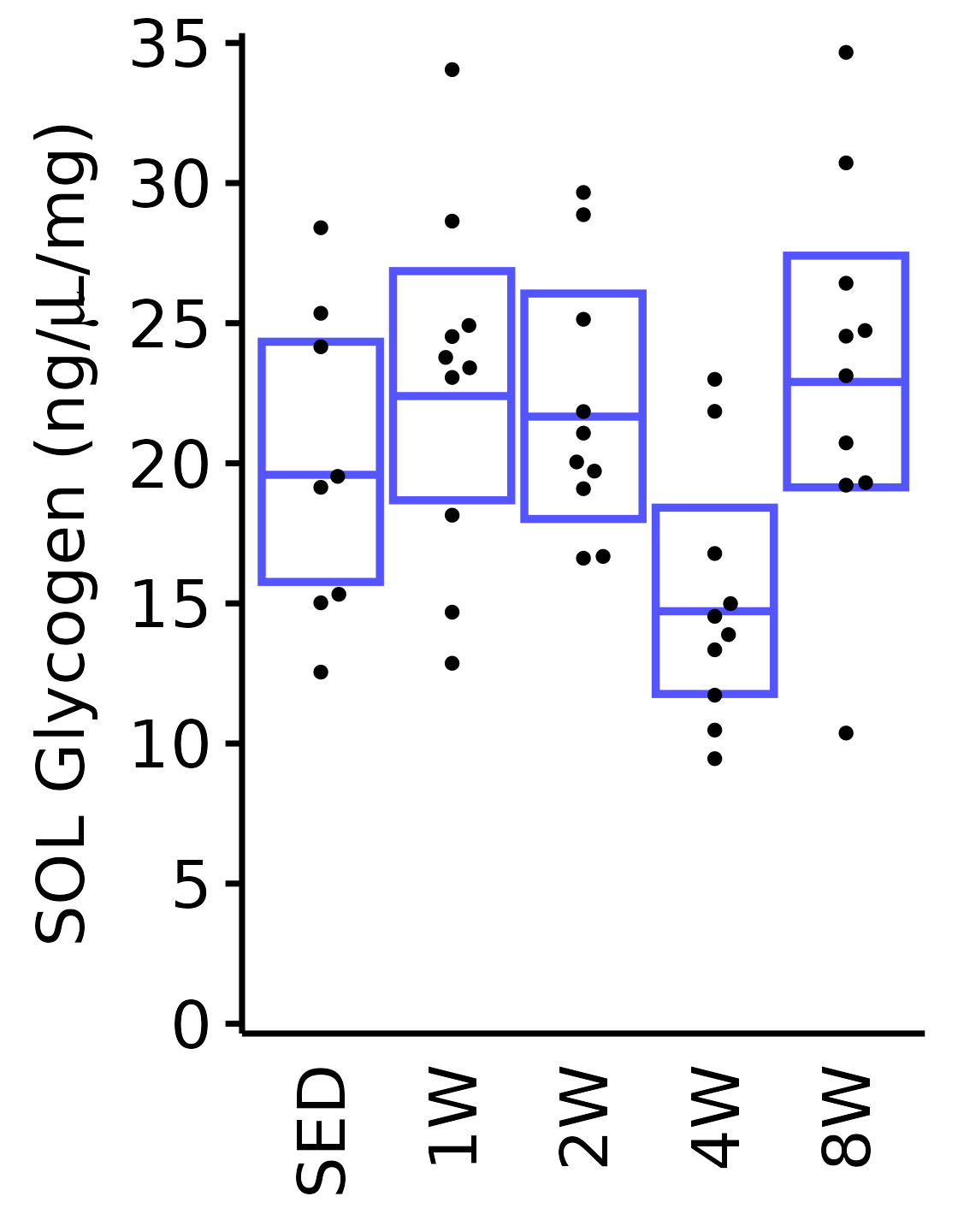
Citrate Synthase
Muscle citrate synthase concentration (U citrate synthase/\(\mu\)g protein \(\times 10^3\)).
LG
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "LG"),
sex = "Female", age = "6M", y_position = 300) +
scale_y_continuous(name = TeX("LG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))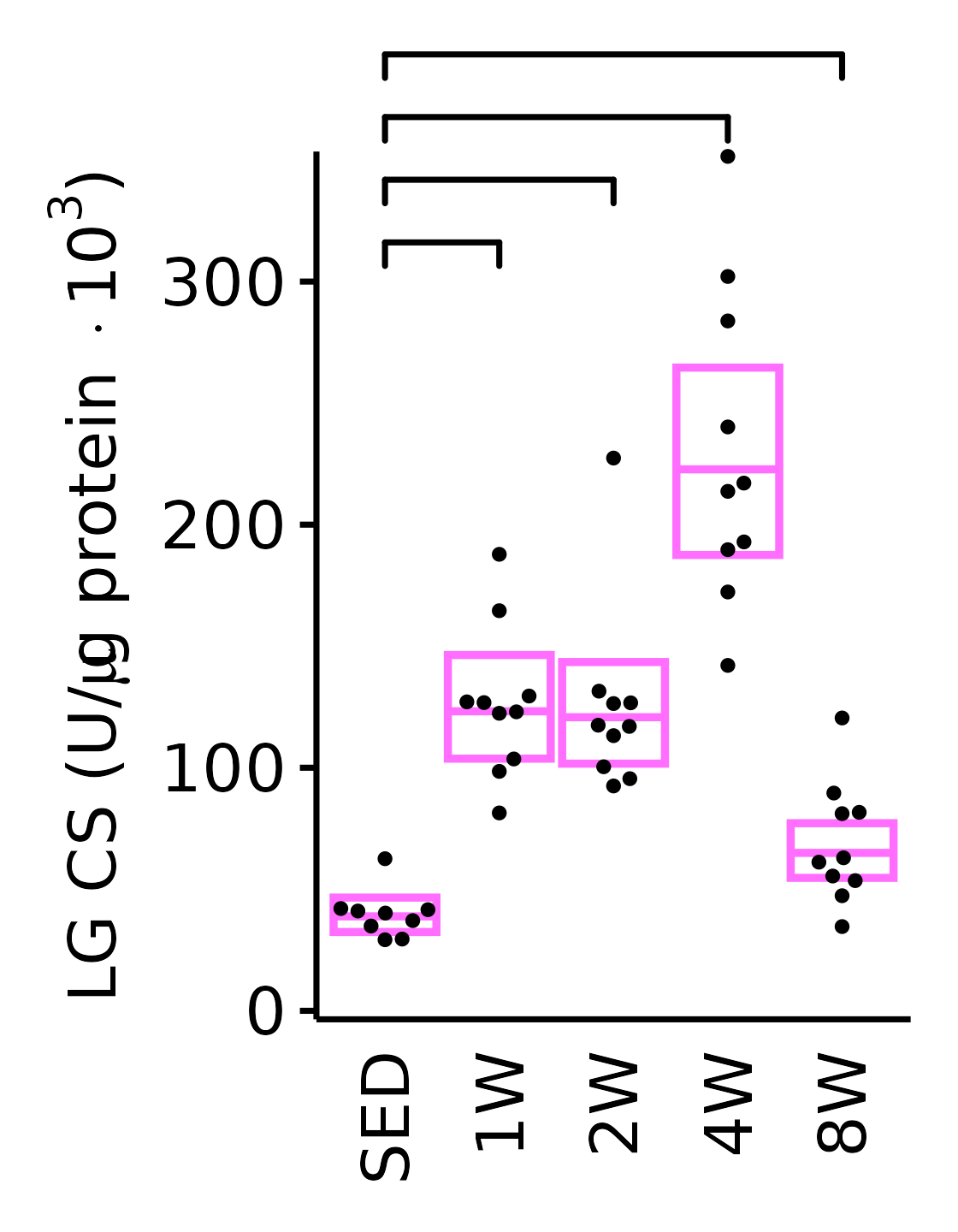
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "LG"),
sex = "Male", age = "6M", y_position = 460) +
scale_y_continuous(name = TeX("LG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))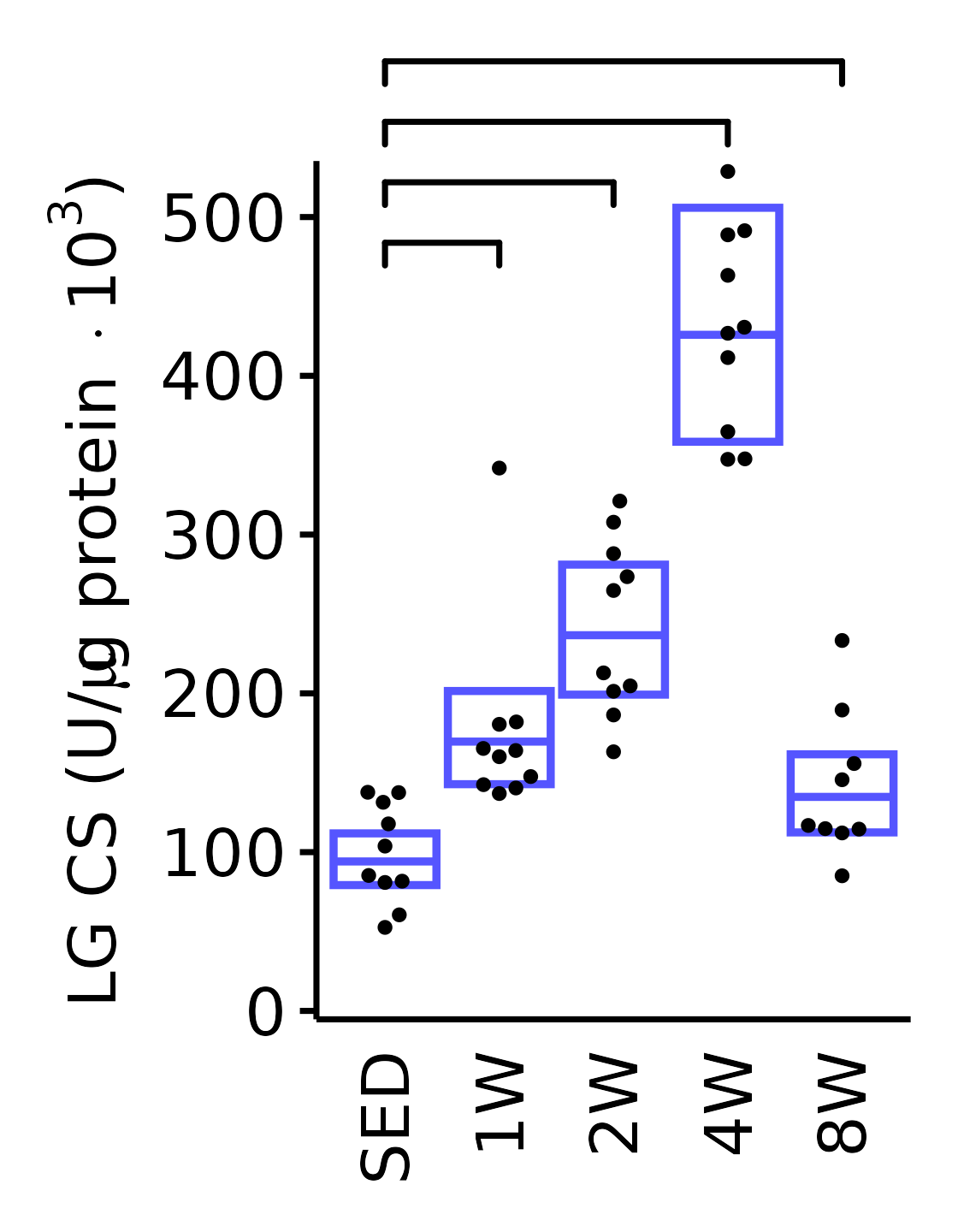
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "LG"),
sex = "Female", age = "18M", y_position = 260) +
scale_y_continuous(name = TeX("LG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))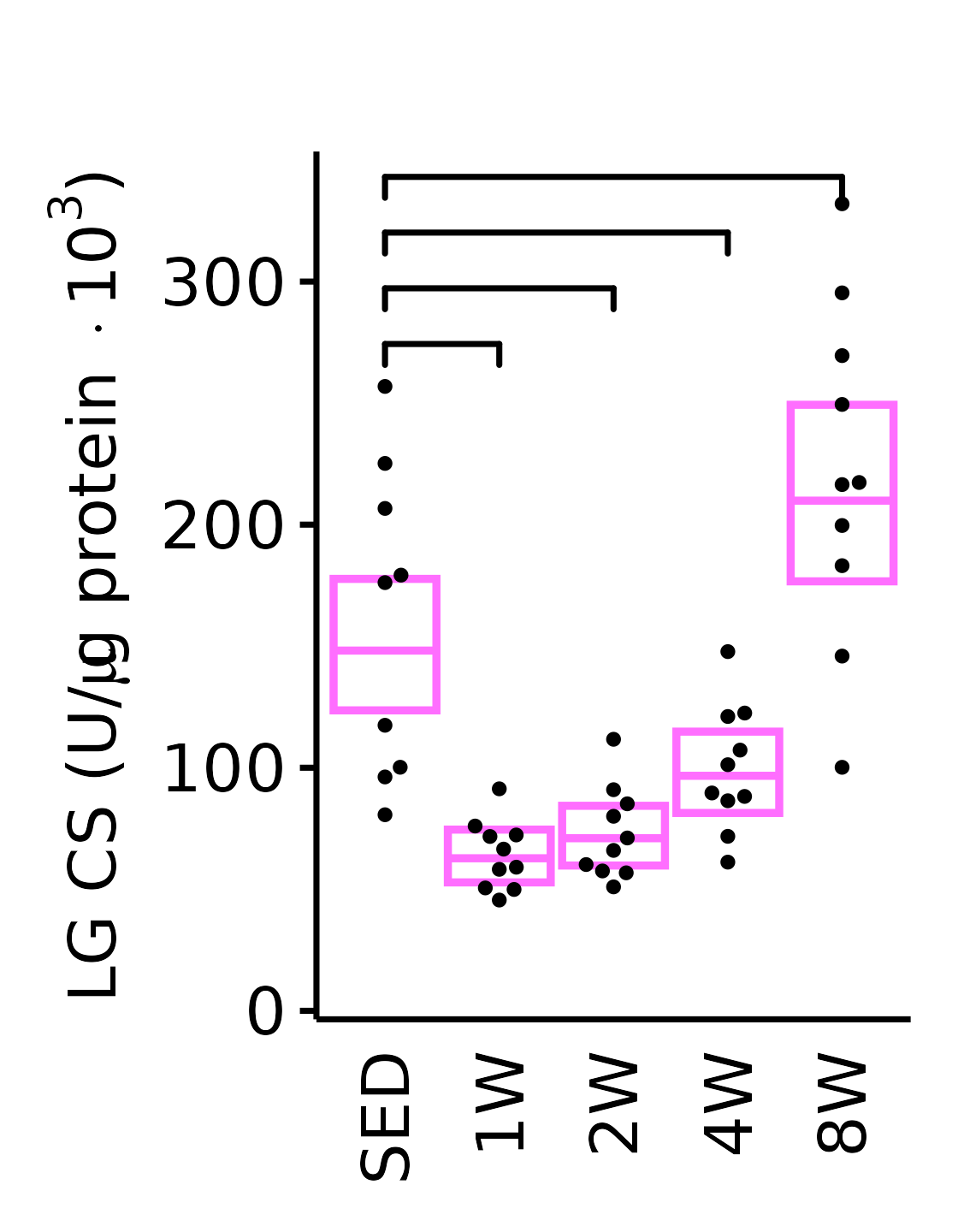
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "LG"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("LG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))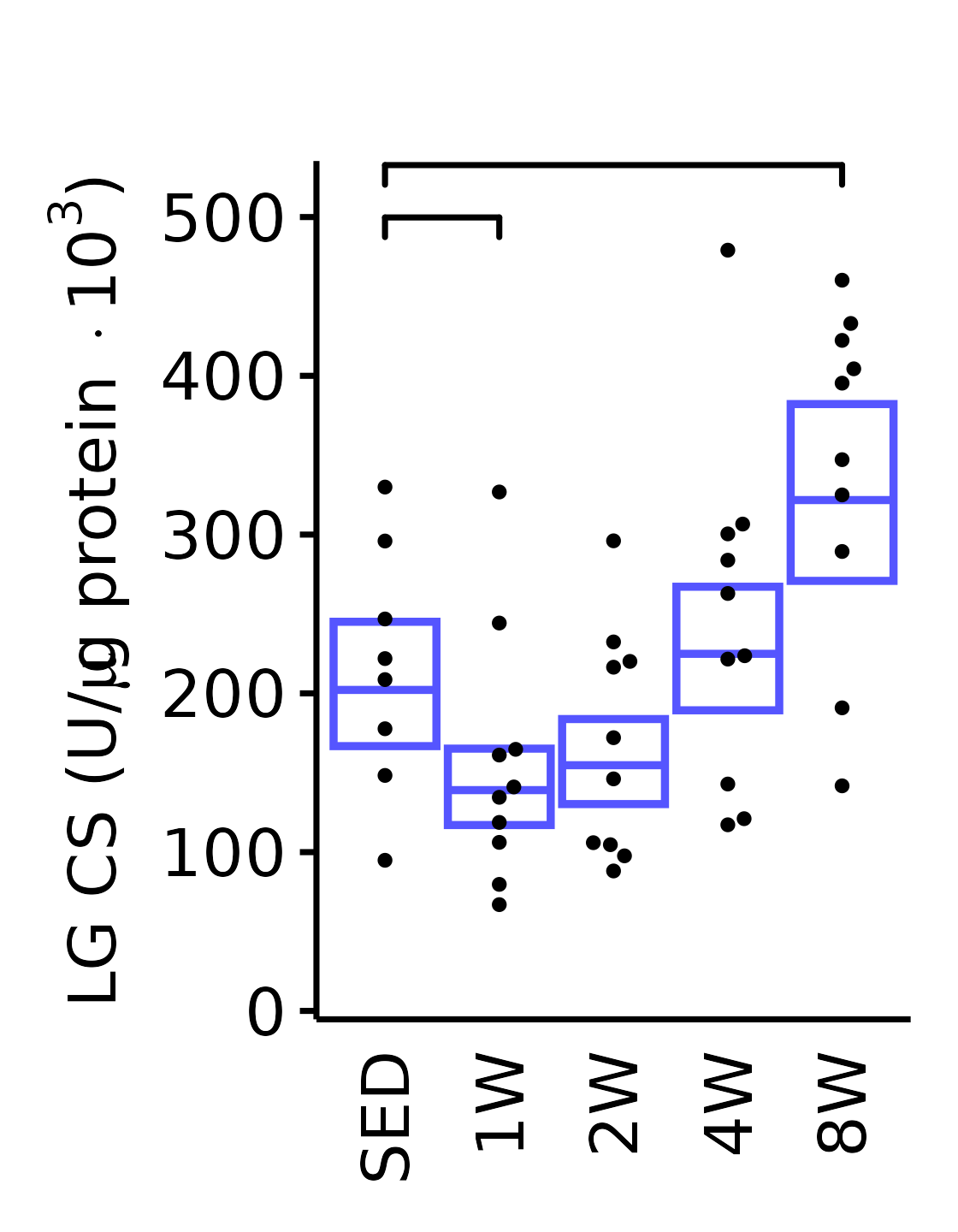
MG
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "MG"),
sex = "Female", age = "6M", y_position = 275) +
scale_y_continuous(name = TeX("MG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))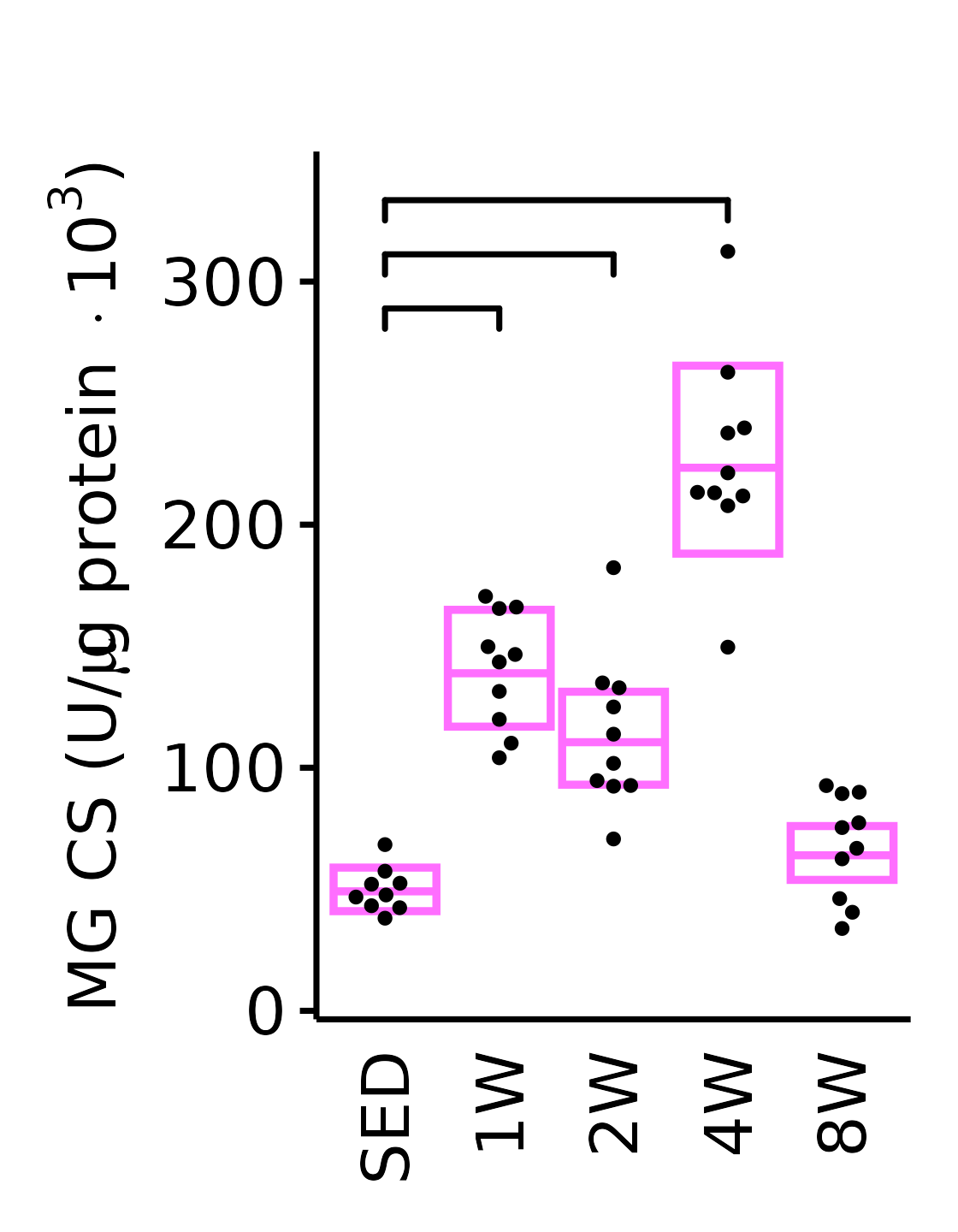
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "MG"),
sex = "Male", age = "6M", y_position = 450) +
scale_y_continuous(name = TeX("MG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))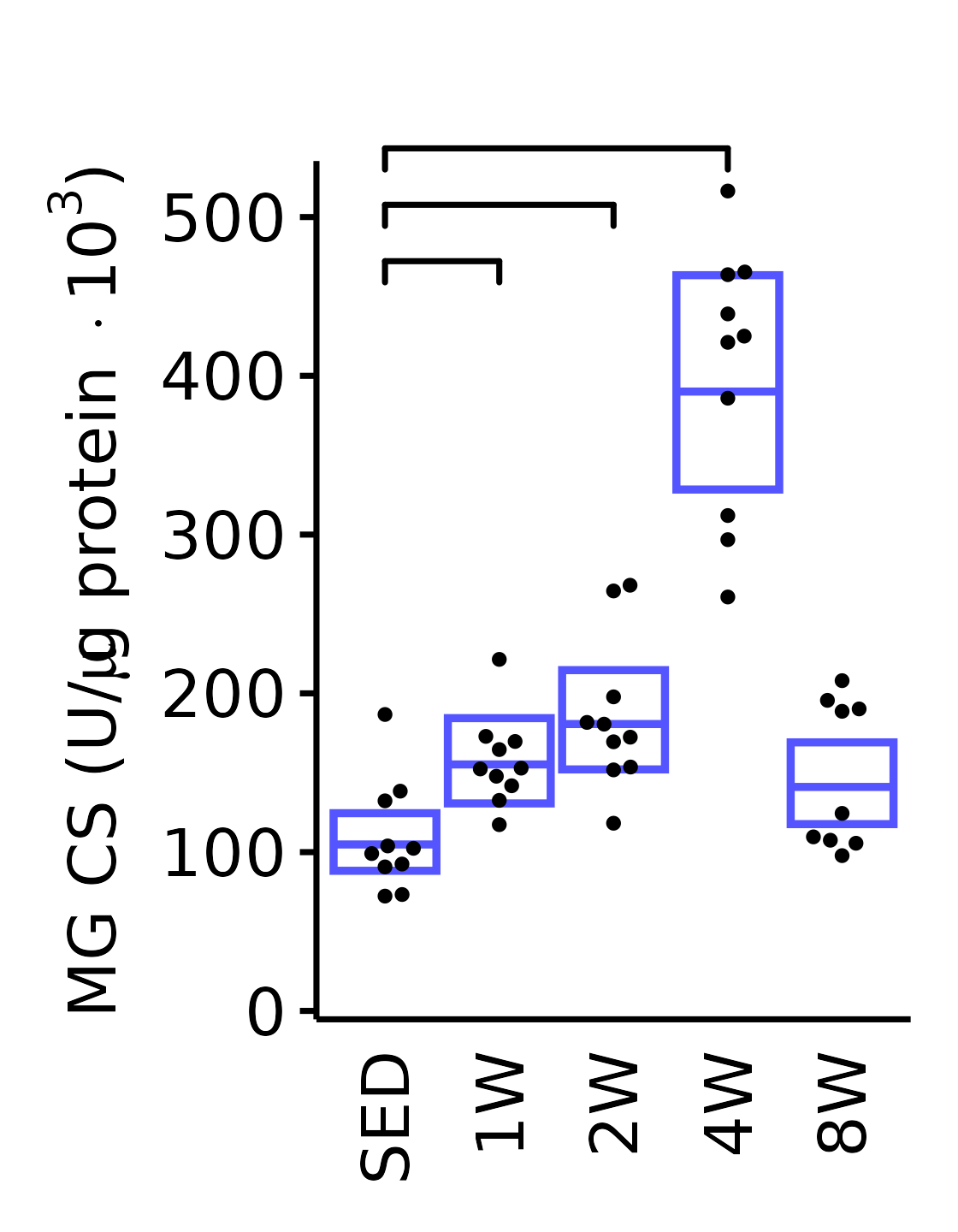
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "MG"),
sex = "Female", age = "18M", y_position = 240) +
scale_y_continuous(name = TeX("MG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))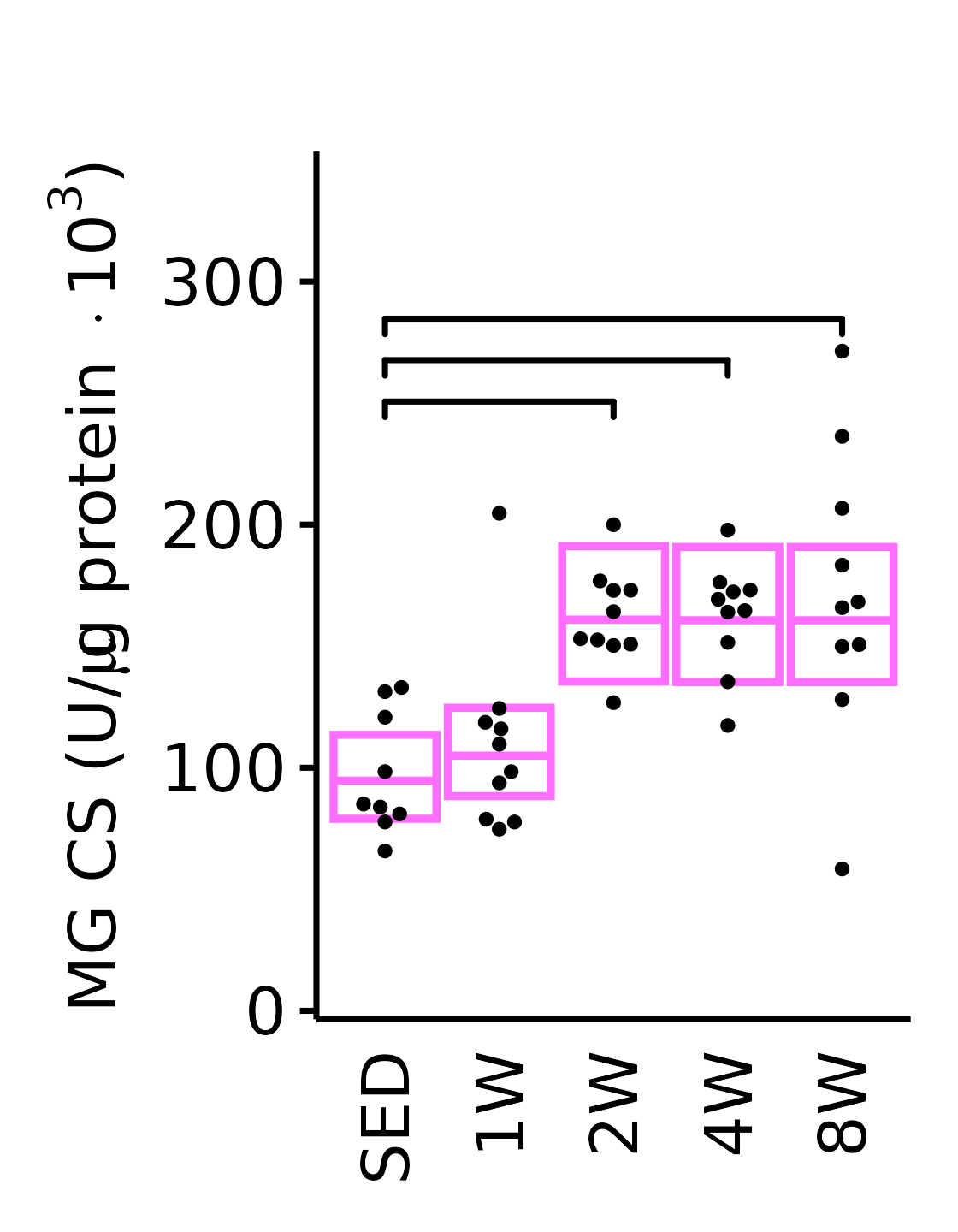
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "MG"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("MG CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))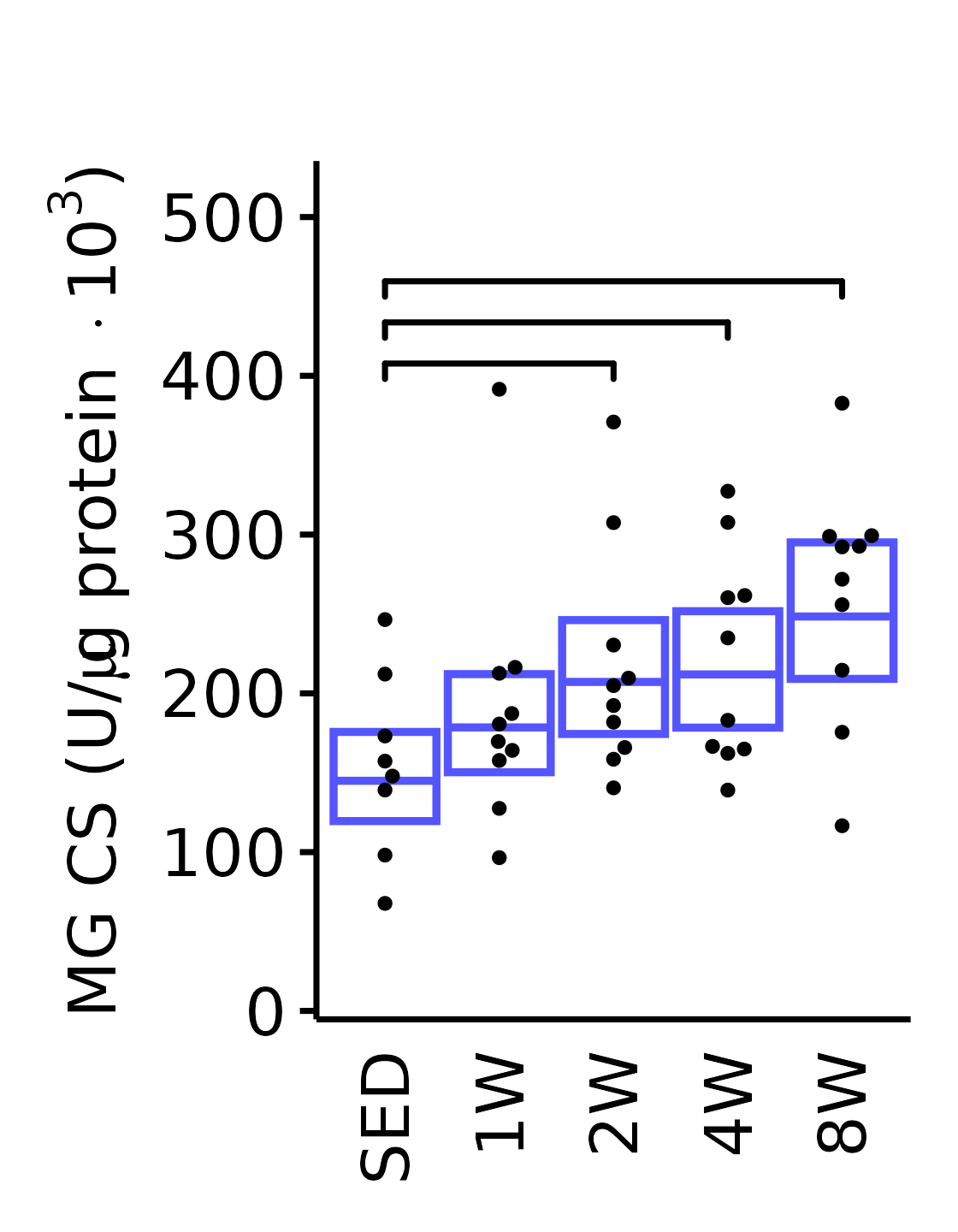
PL
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "PL"),
sex = "Female", age = "6M", y_position = 210) +
scale_y_continuous(name = TeX("PL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))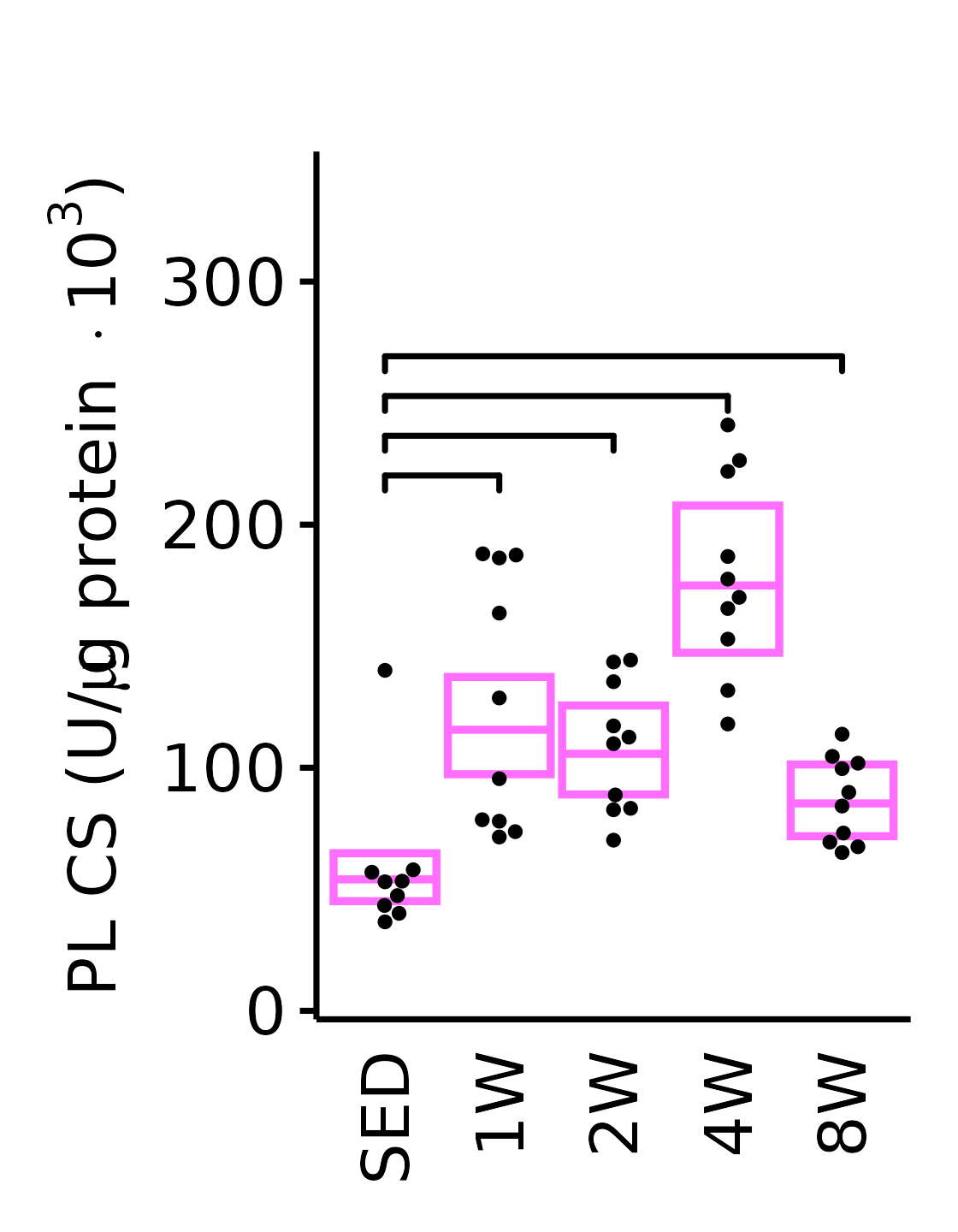
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "PL"),
sex = "Male", age = "6M", y_position = 320) +
scale_y_continuous(name = TeX("PL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))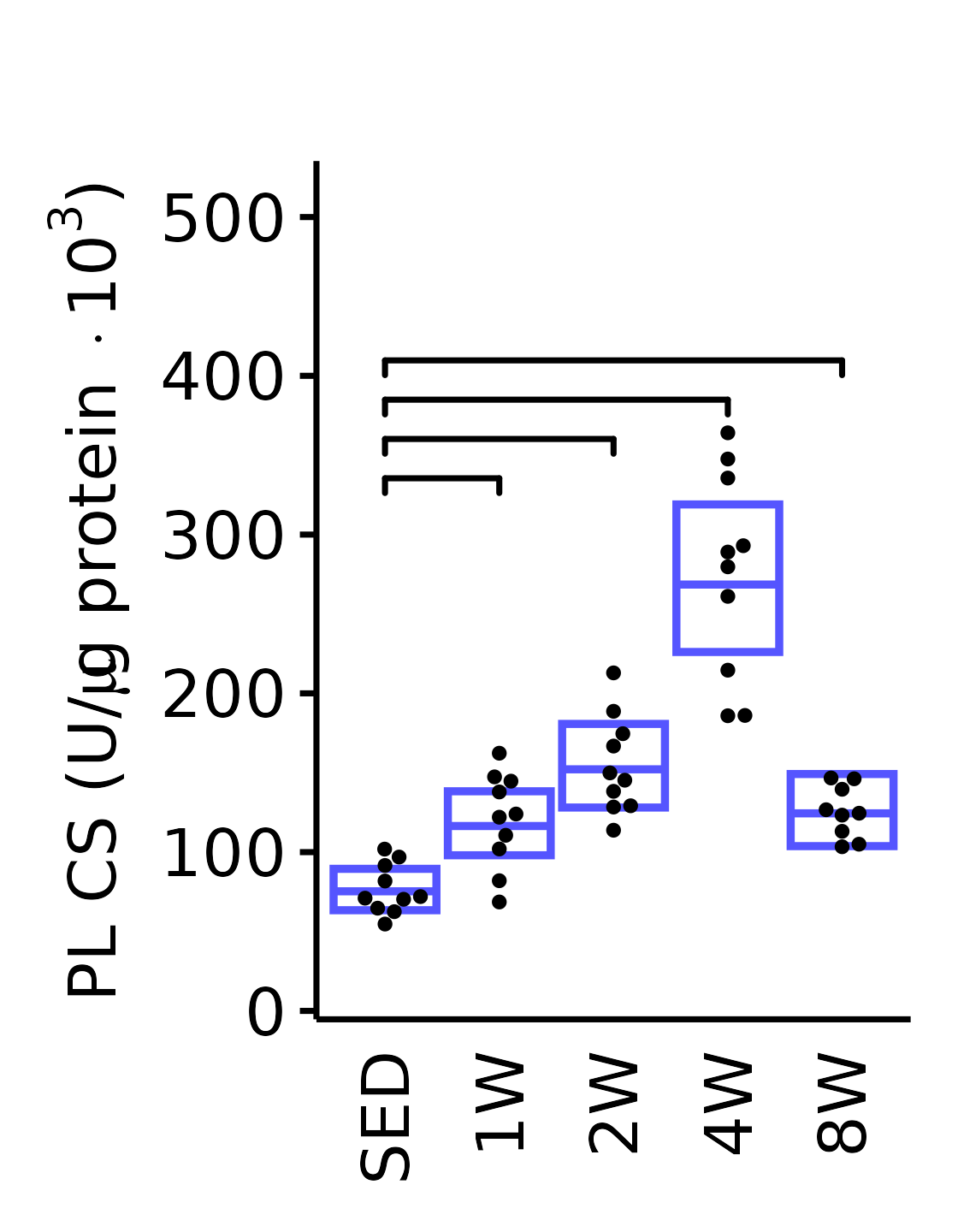
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "PL"),
sex = "Female", age = "18M", y_position = 210) +
scale_y_continuous(name = TeX("PL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))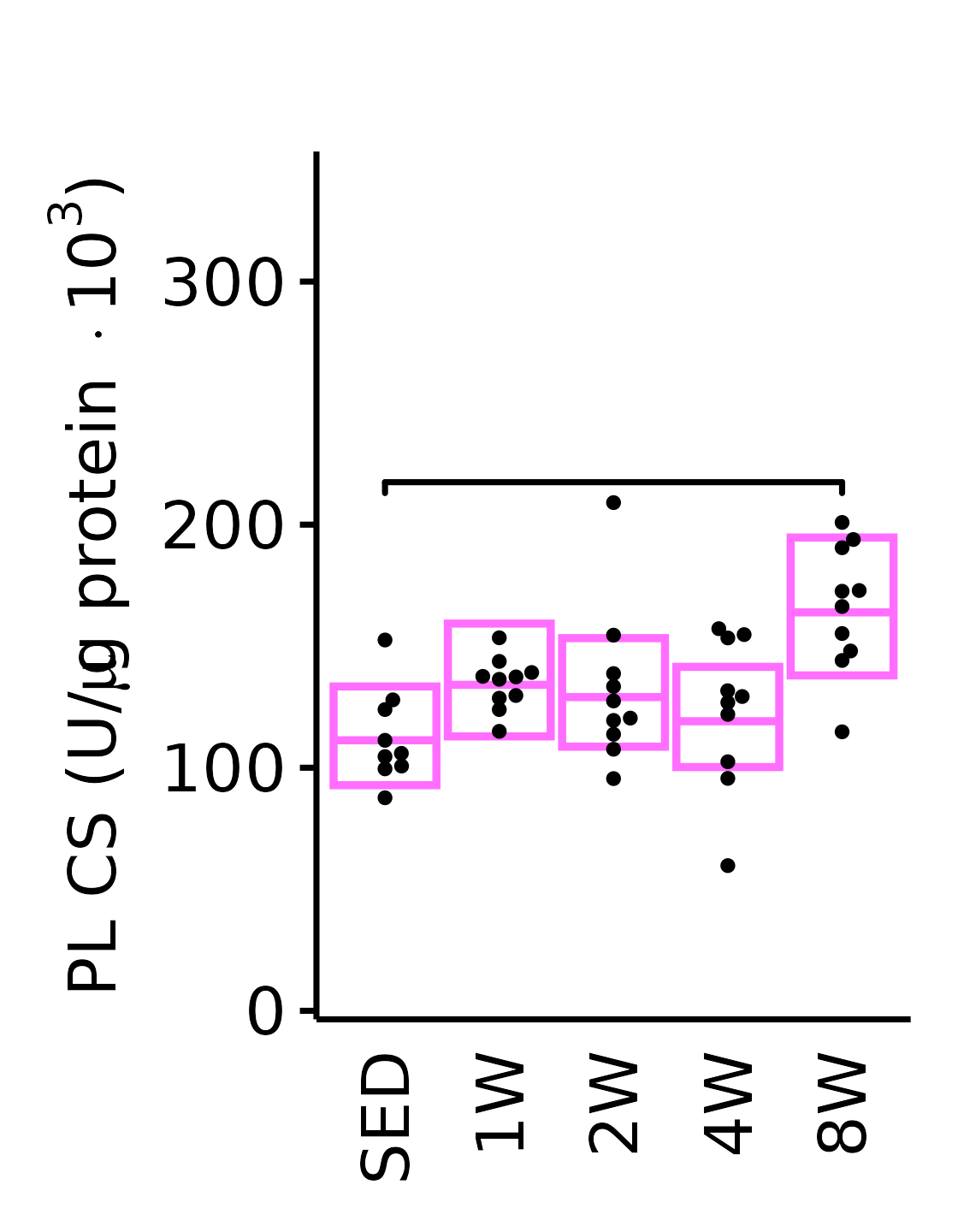
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "PL"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("PL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))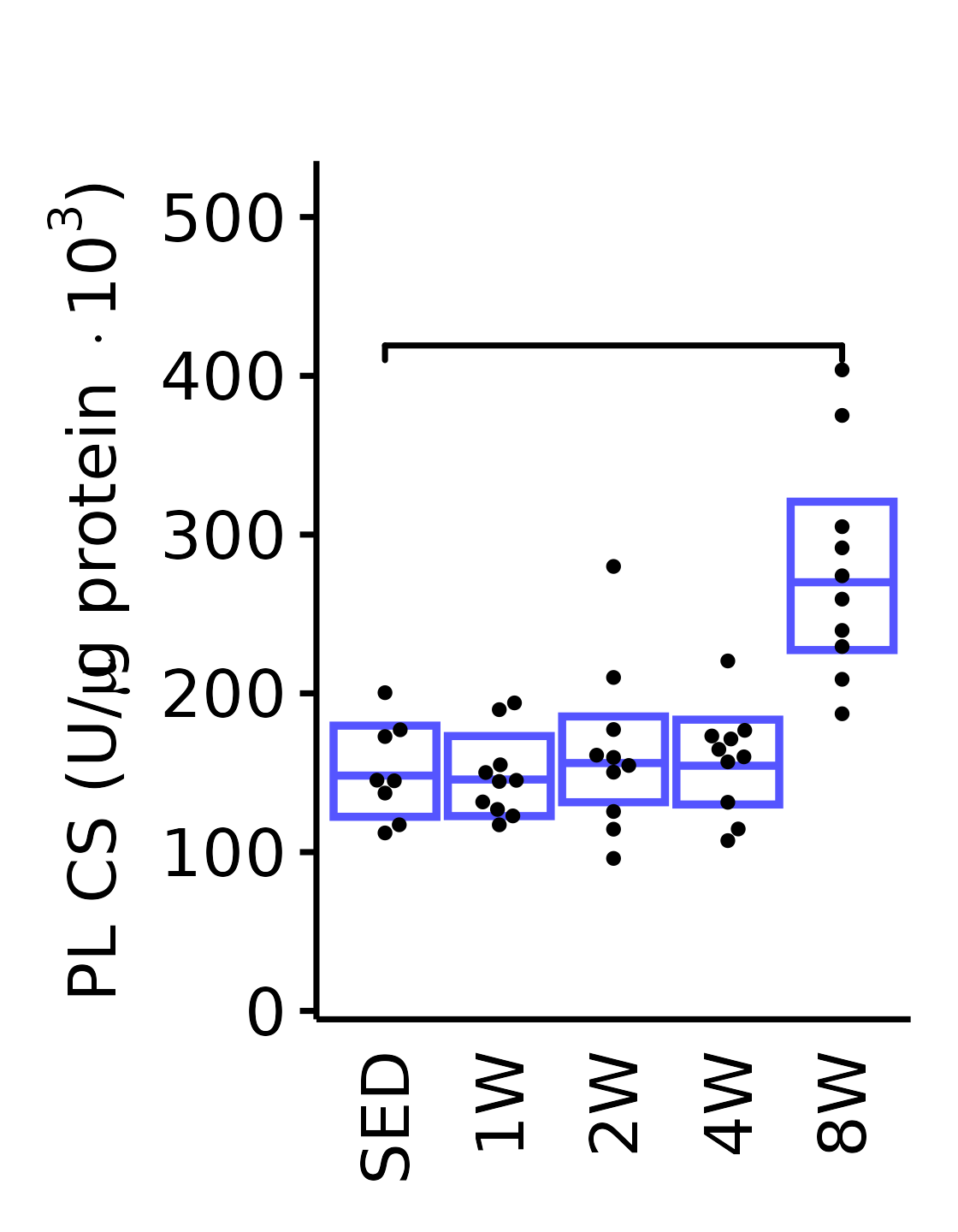
SOL
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "SOL"),
sex = "Female", age = "6M", y_position = 310) +
scale_y_continuous(name = TeX("SOL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))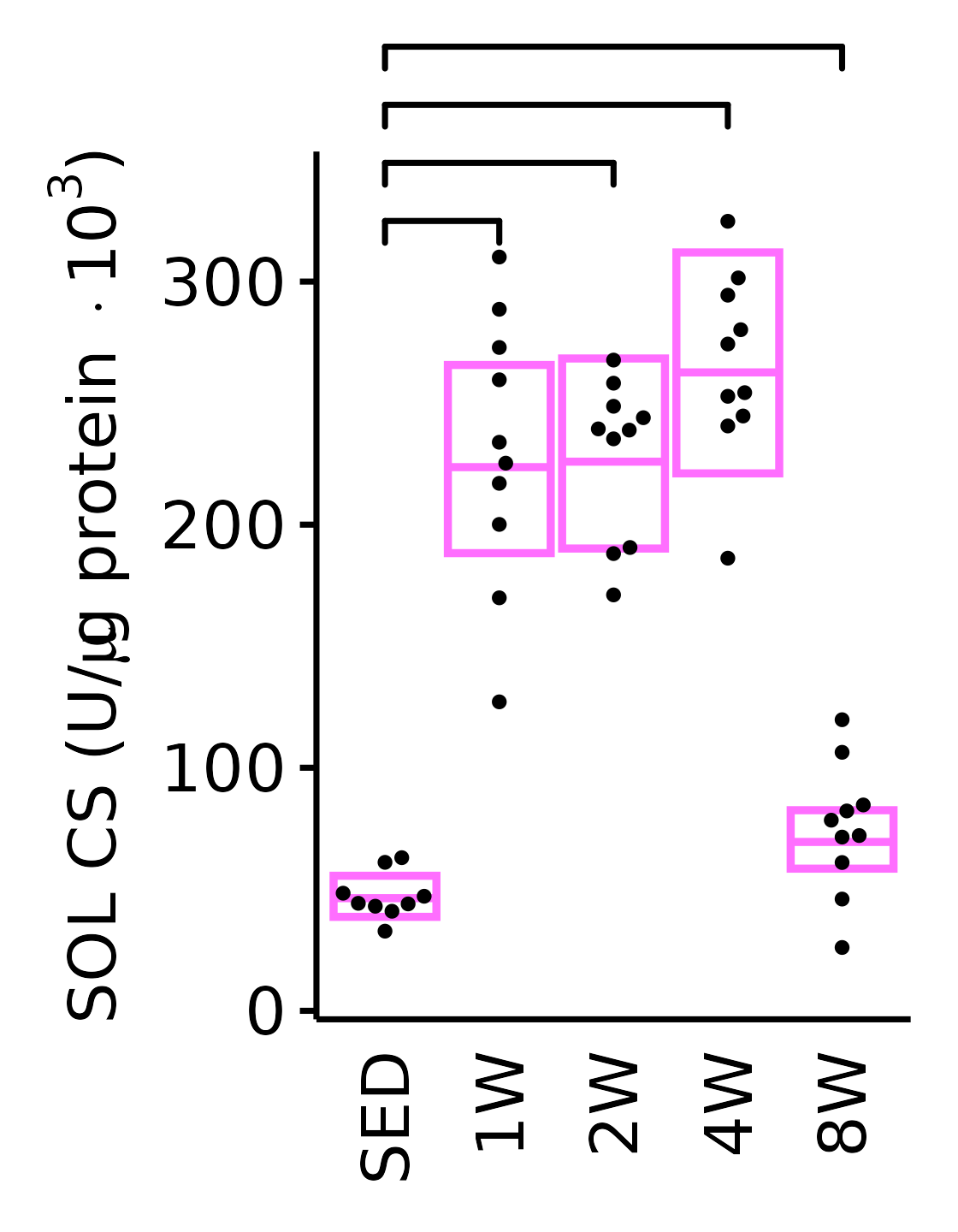
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "SOL"),
sex = "Male", age = "6M", y_position = 450) +
scale_y_continuous(name = TeX("SOL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))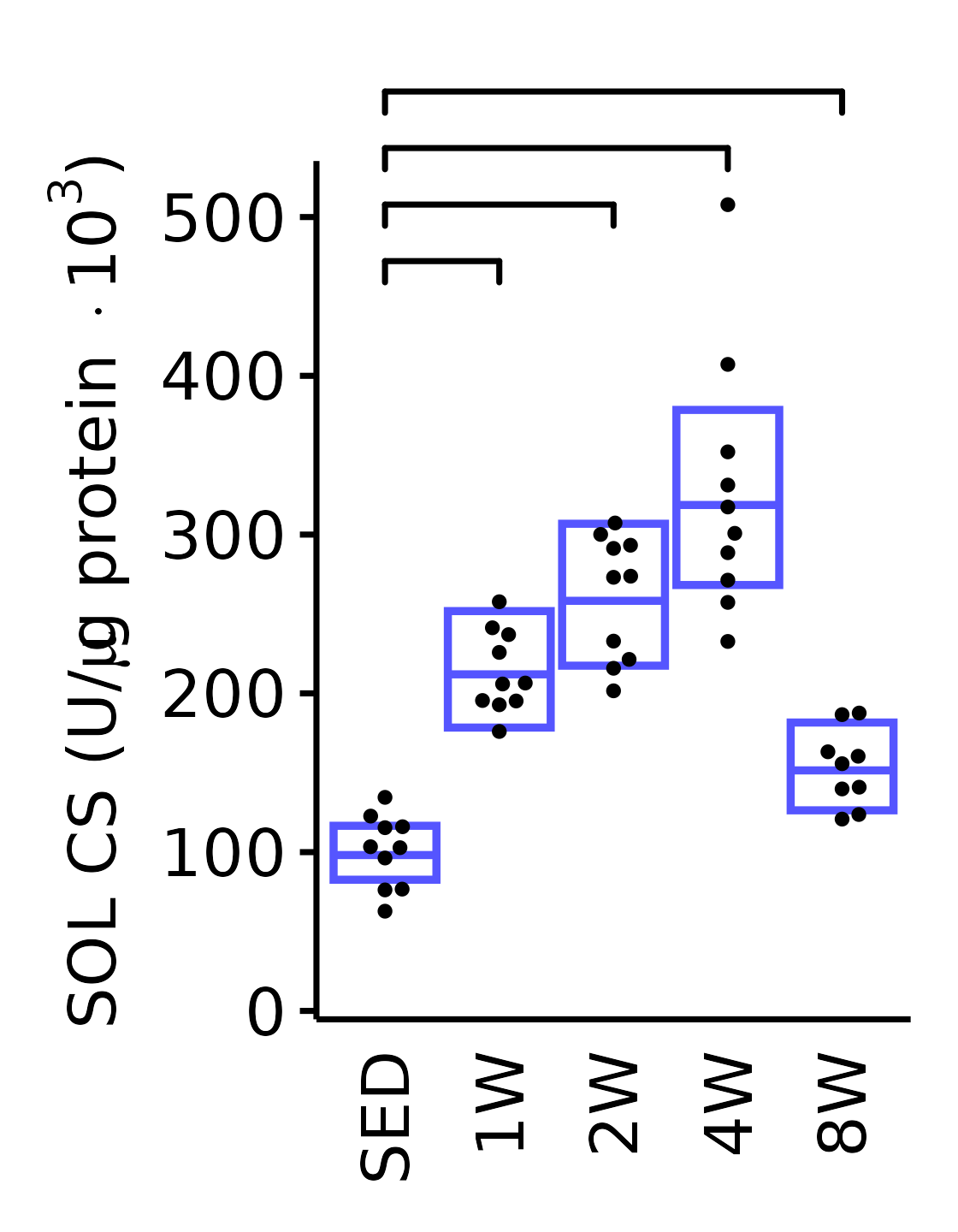
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "SOL"),
sex = "Female", age = "18M", y_position = 230) +
scale_y_continuous(name = TeX("SOL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 350, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 350), clip = "off") +
theme(plot.margin = unit(c(16, 5, 5, 5), units = "pt"))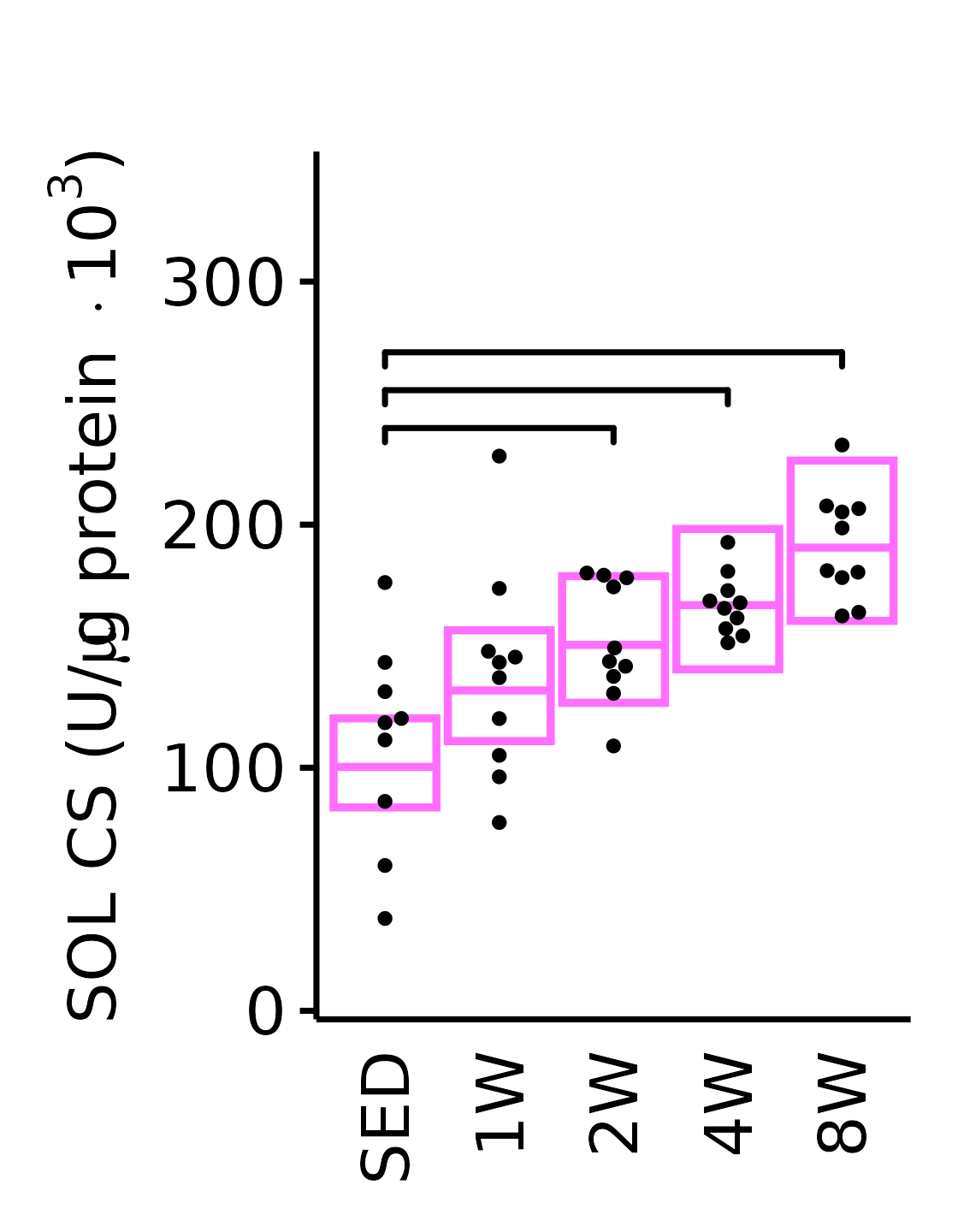
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "citrate_synthase",
conf = filter(conf_df, response == "Citrate Synthase",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Citrate Synthase",
muscle == "SOL"),
sex = "Male", age = "18M") +
scale_y_continuous(name = TeX("SOL CS (U/$\\mu$g protein $\\times 10^3$)"),
breaks = seq(0, 500, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(0, 530), clip = "off") +
theme(plot.margin = unit(c(17, 5, 5, 5), units = "pt"))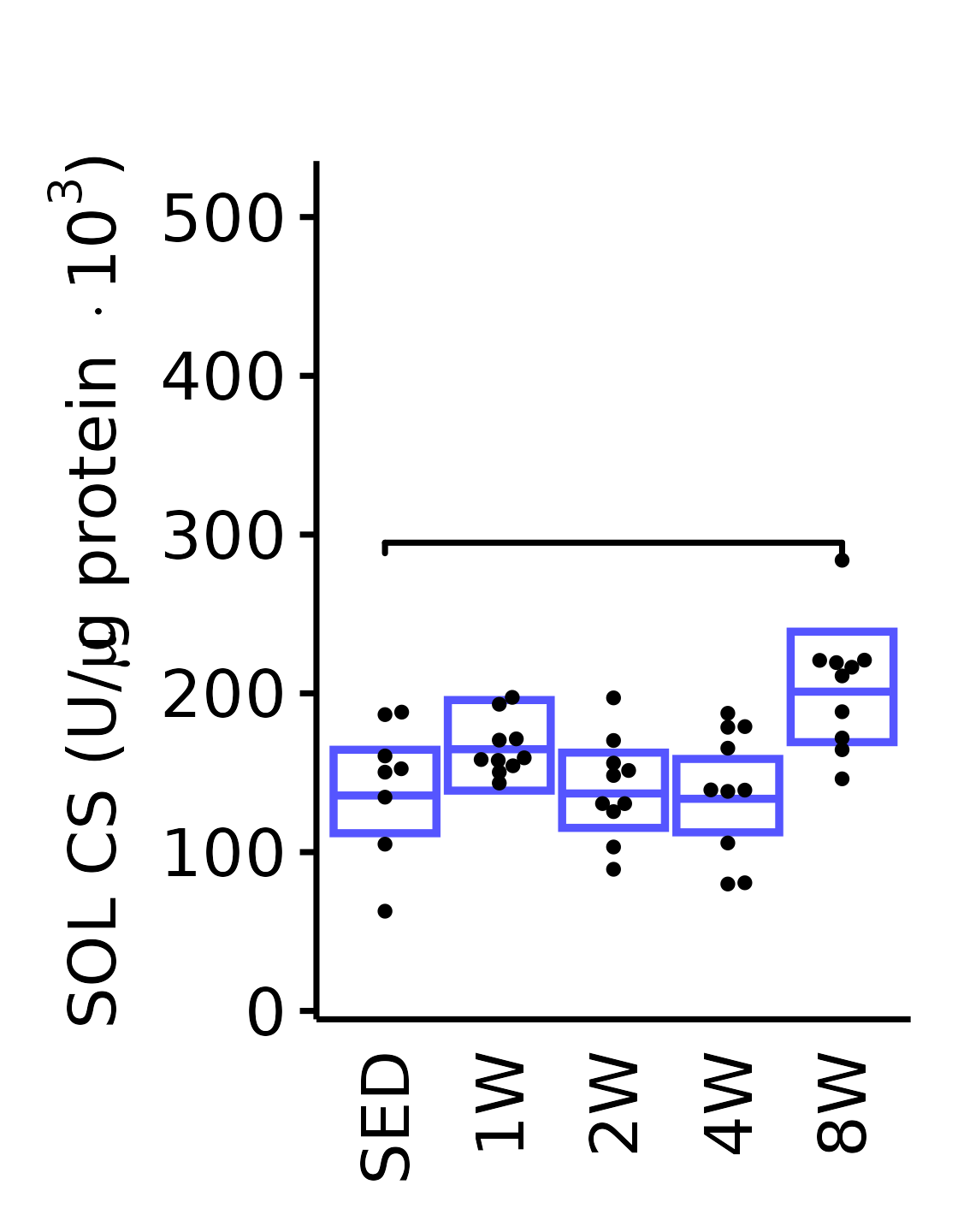
Terminal Muscle Mass
Mass (mg) of dissected muscles.
LG
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "LG"),
sex = "Female", age = "6M", y_position = 620) +
scale_y_continuous(name = "LG Terminal Mass (mg)",
breaks = seq(500, 700, 50),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(485, 700), clip = "off")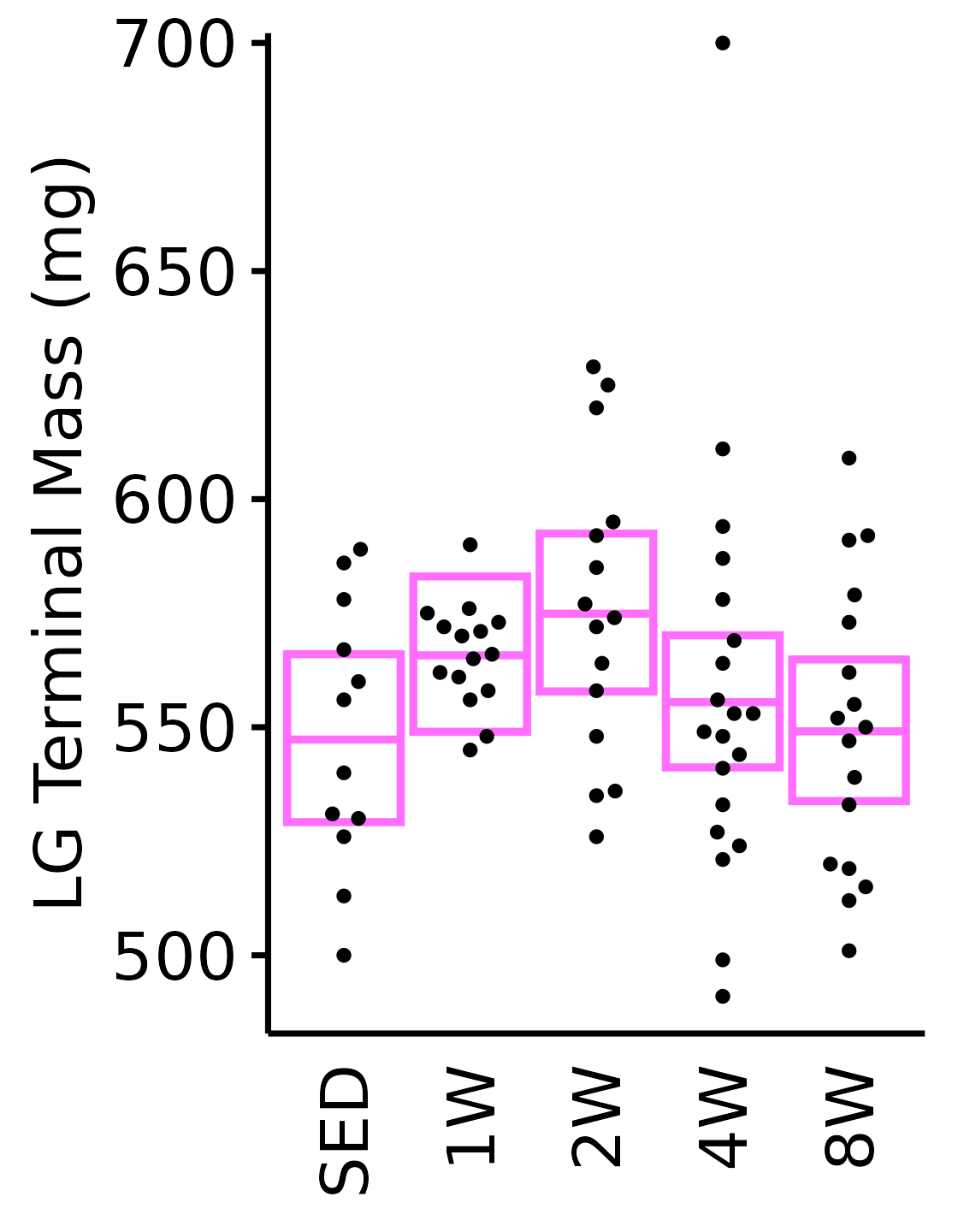
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "LG"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "LG Terminal Mass (mg)",
breaks = seq(700, 1000, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(650, 1060), clip = "off")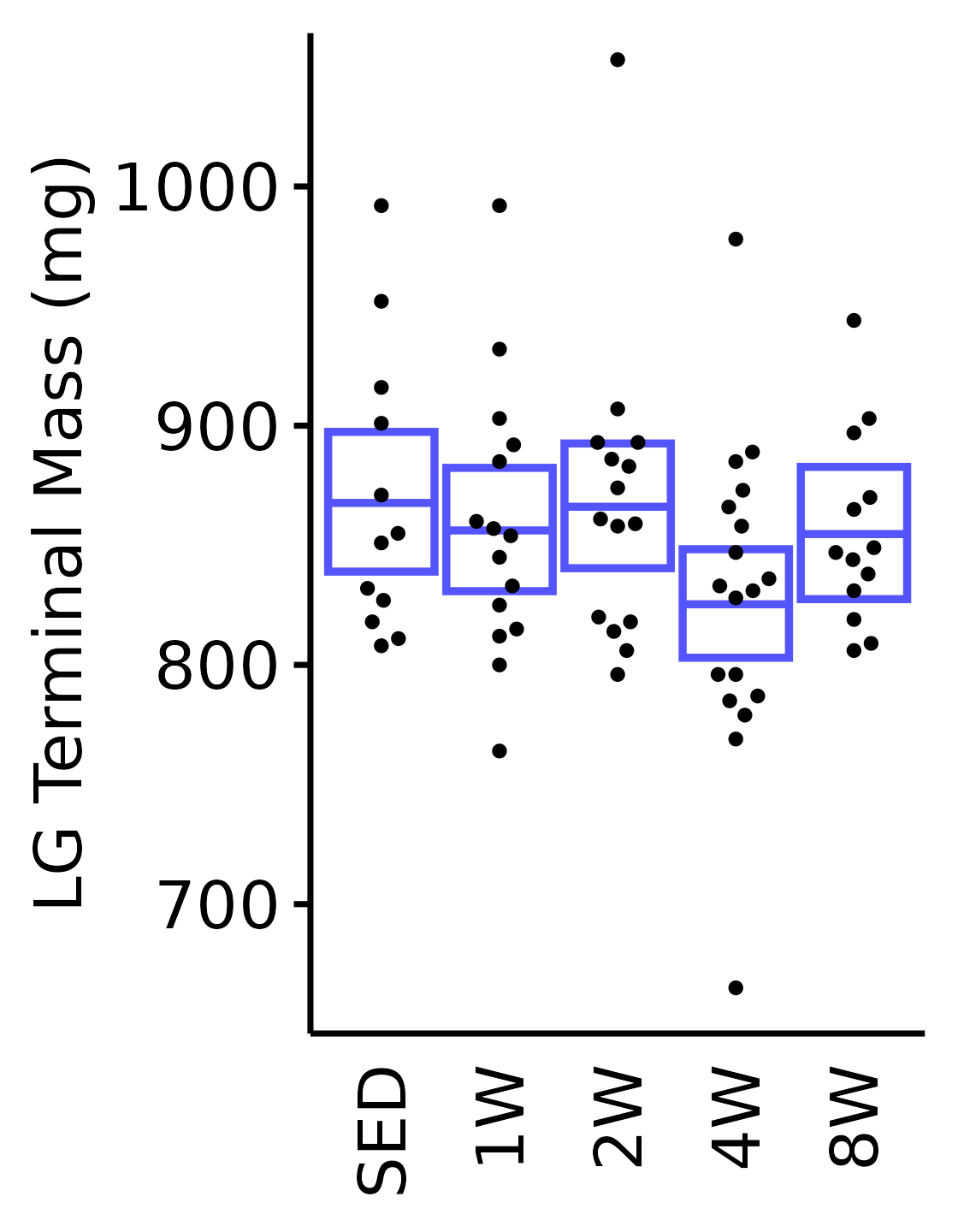
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "LG"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "LG Terminal Mass (mg)",
breaks = seq(500, 700, 50),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(485, 700), clip = "off")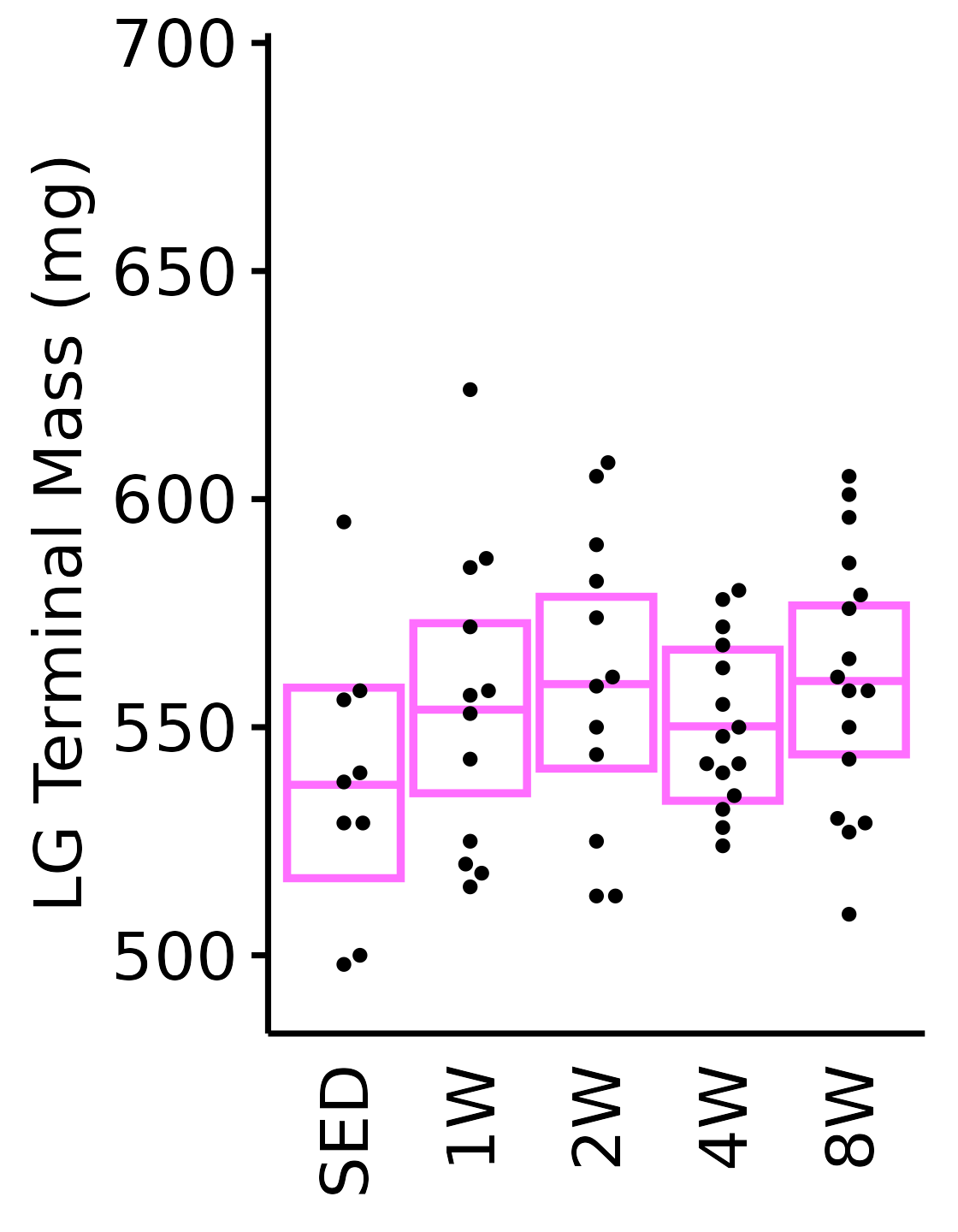
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "LG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "LG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "LG"),
sex = "Male", age = "18M", y_position = 940) +
scale_y_continuous(name = "LG Terminal Mass (mg)",
breaks = seq(700, 1000, 100),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(650, 1060), clip = "off")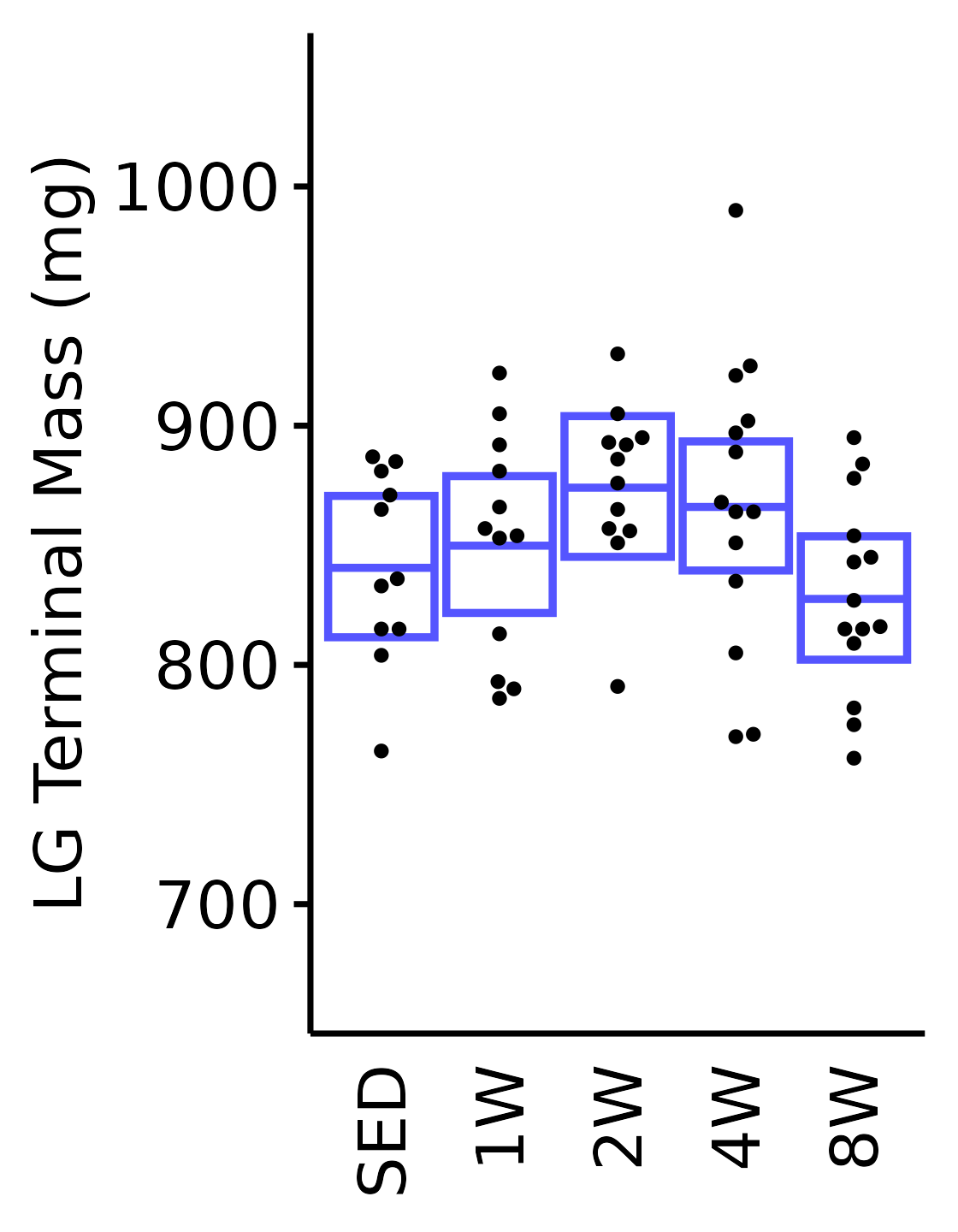
MG
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "MG"),
sex = "Female", age = "6M") +
scale_y_continuous(name = "MG Terminal Mass (mg)",
breaks = seq(320, 470, 30),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(315, 472), clip = "off") +
theme(plot.margin = unit(c(10, 5, 5, 5), units = "pt"))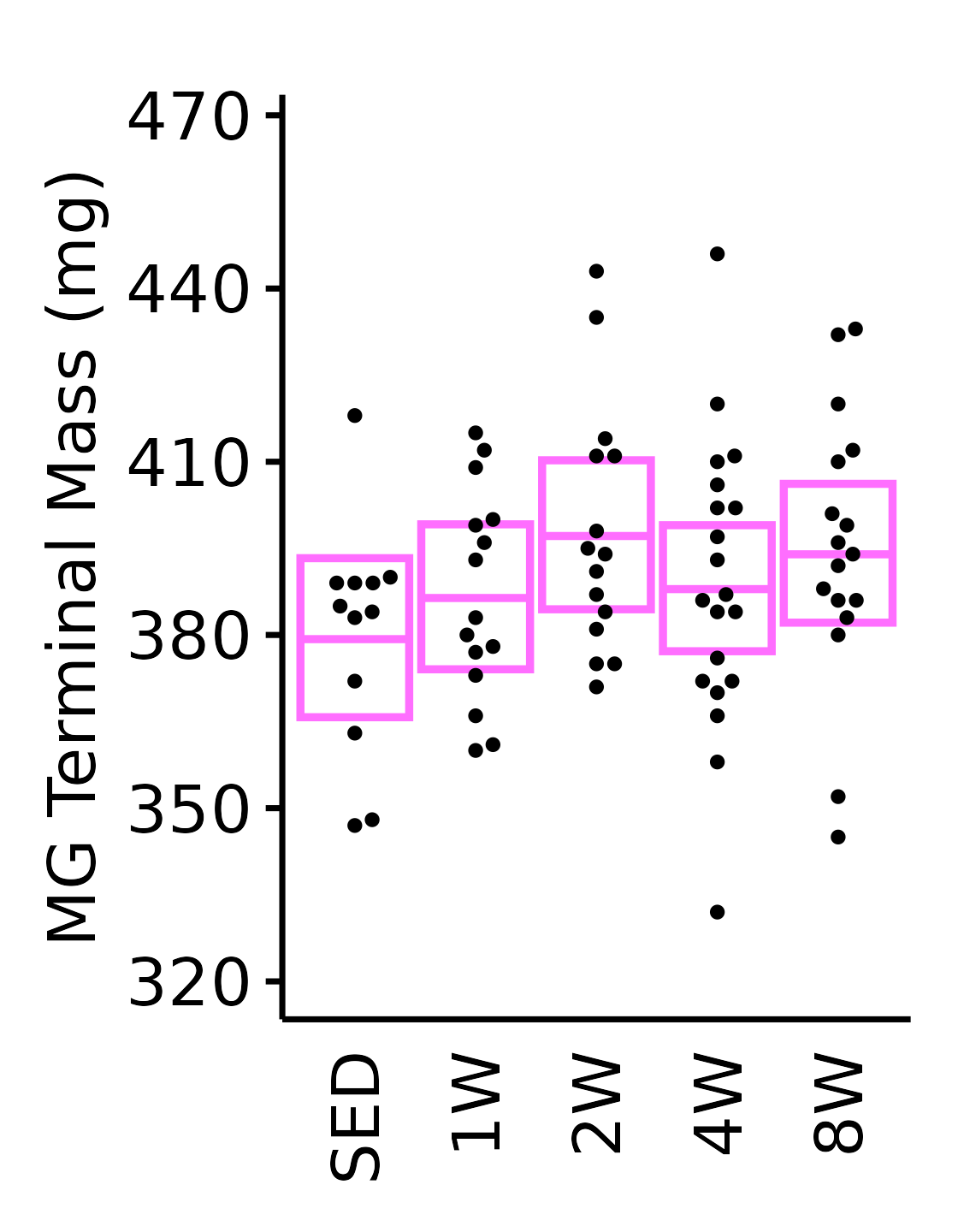
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "MG"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "MG Terminal Mass (mg)",
breaks = seq(500, 750, 50),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(500, 770), clip = "off")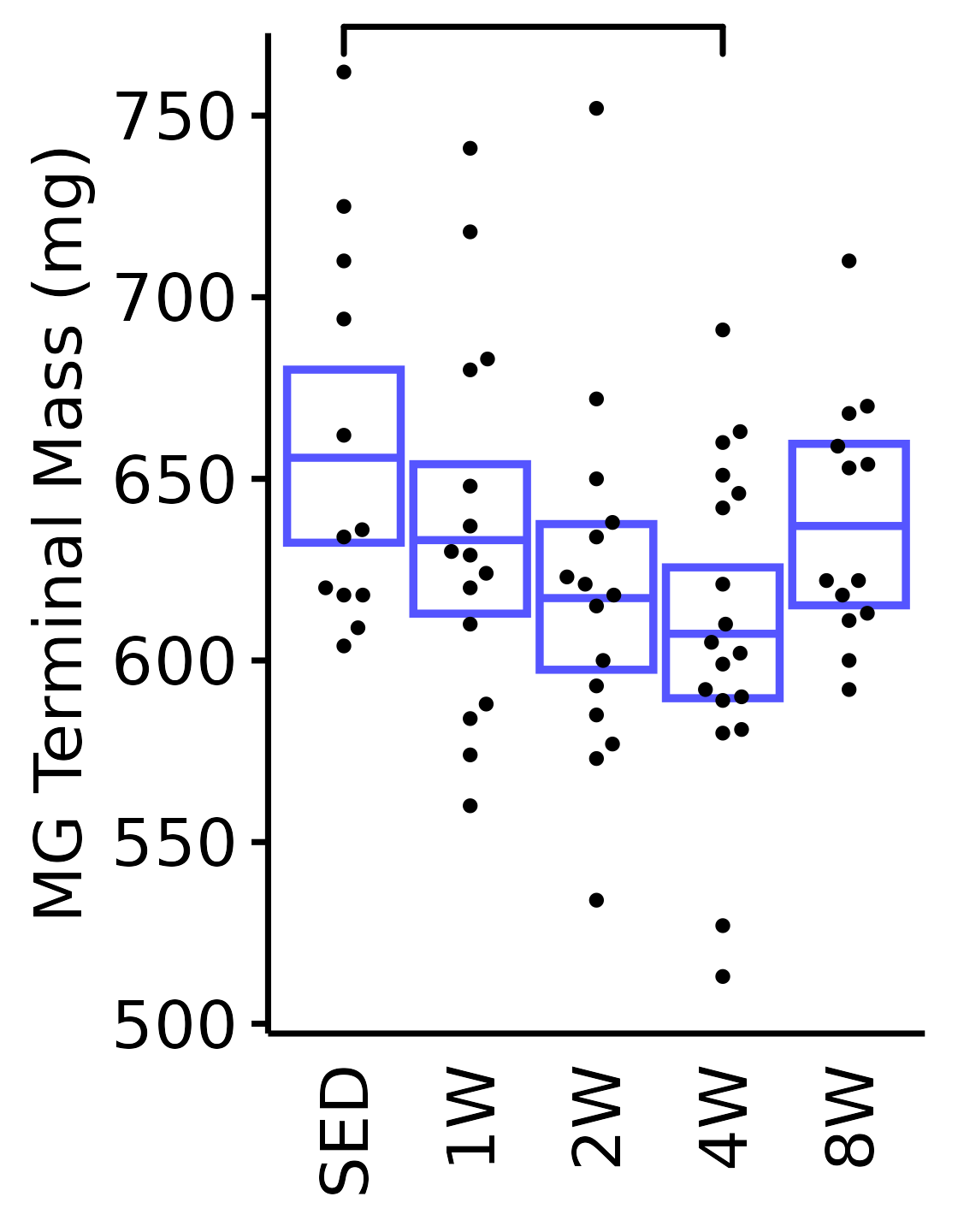
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "MG"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "MG Terminal Mass (mg)",
breaks = seq(320, 470, 30),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(315, 472), clip = "off") +
theme(plot.margin = unit(c(10, 5, 5, 5), units = "pt"))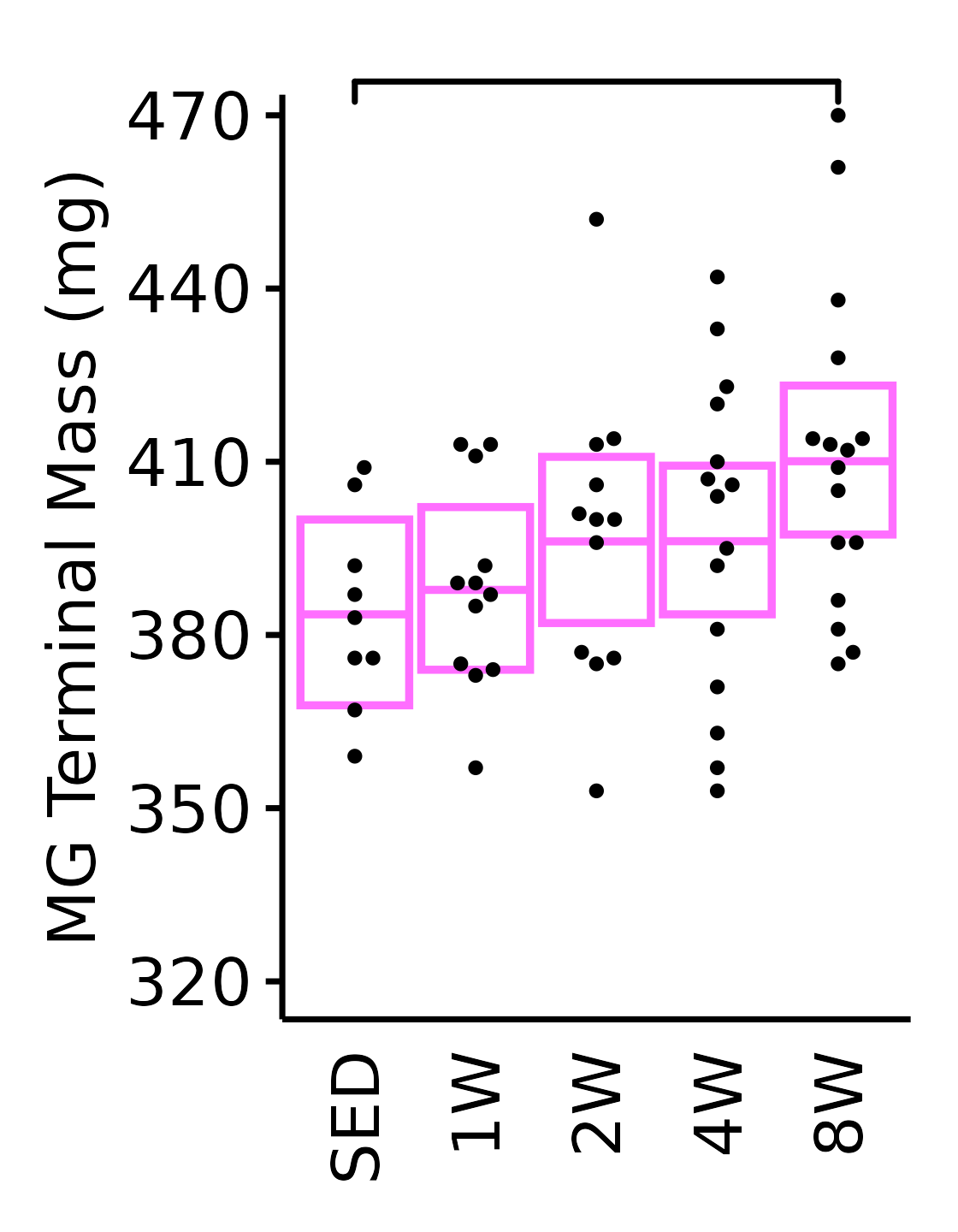
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "MG"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "MG"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "MG"),
sex = "Male", age = "18M") +
scale_y_continuous(name = "MG Terminal Mass (mg)",
breaks = seq(500, 750, 50),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(500, 770), clip = "off")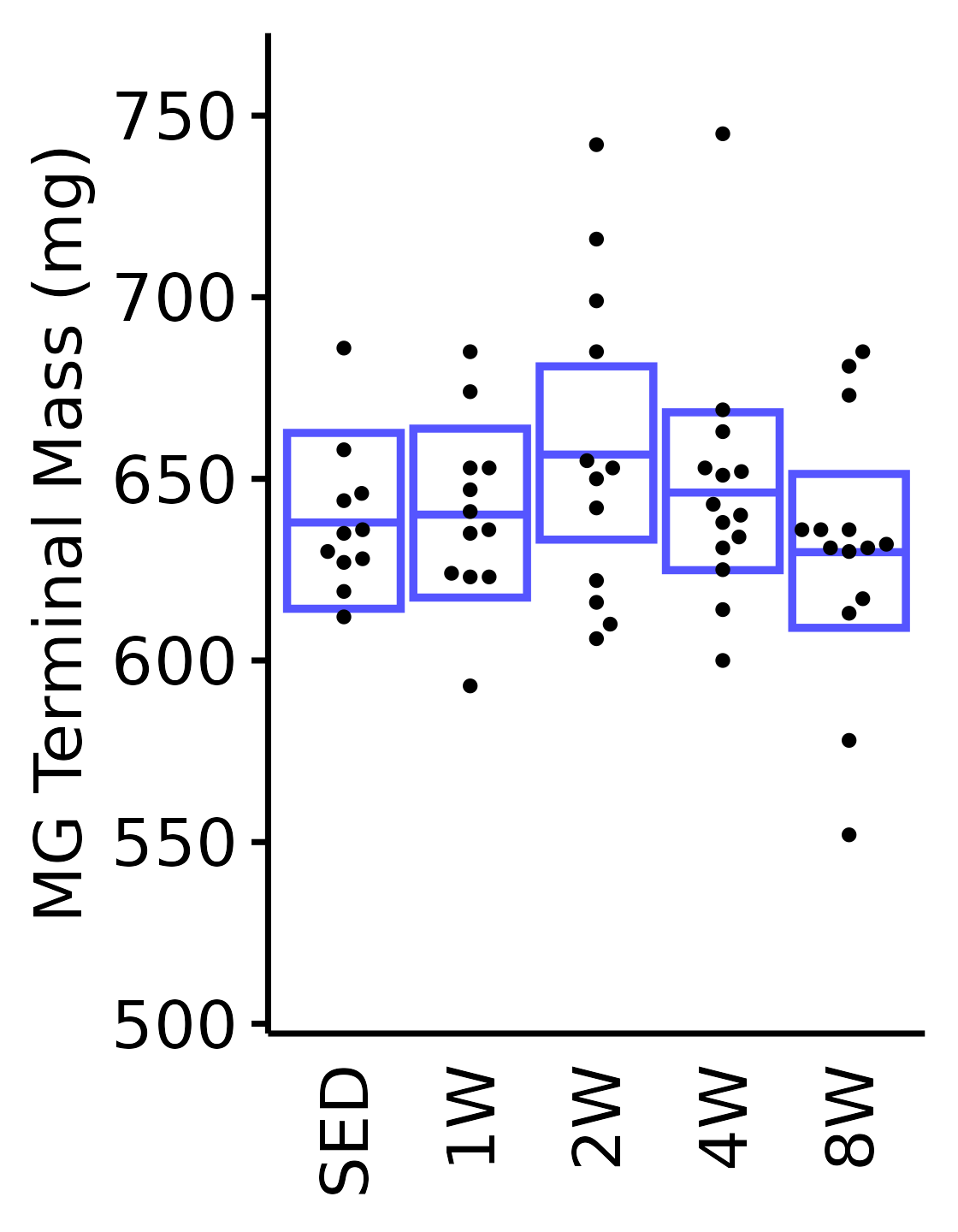
PL
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "PL"),
sex = "Female", age = "6M", y_position = 218) +
scale_y_continuous(name = "PL Terminal Mass (mg)",
breaks = seq(160, 220, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(160, 225), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))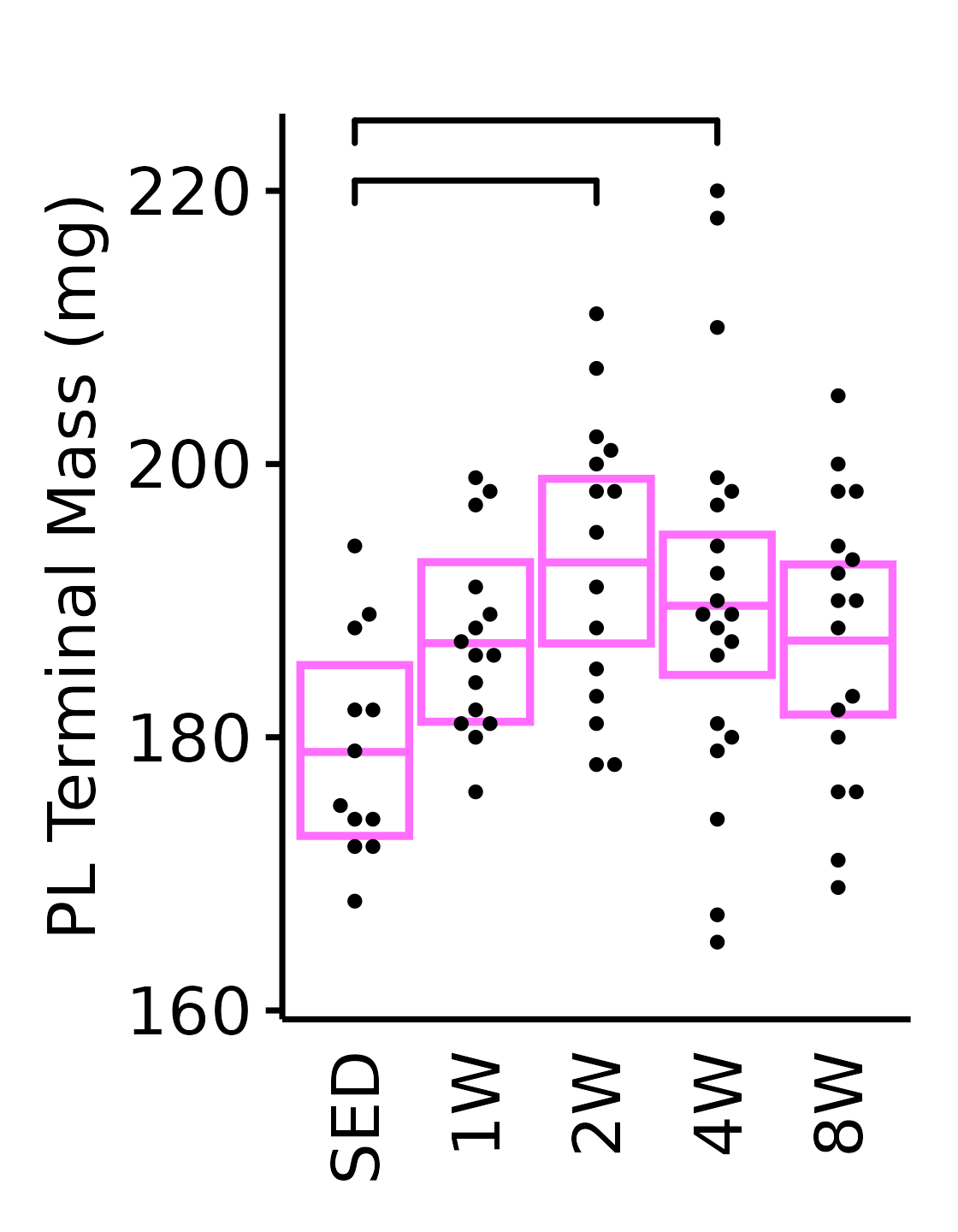
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "PL"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "PL Terminal Mass (mg)",
breaks = seq(200, 380, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(200, 380), clip = "off")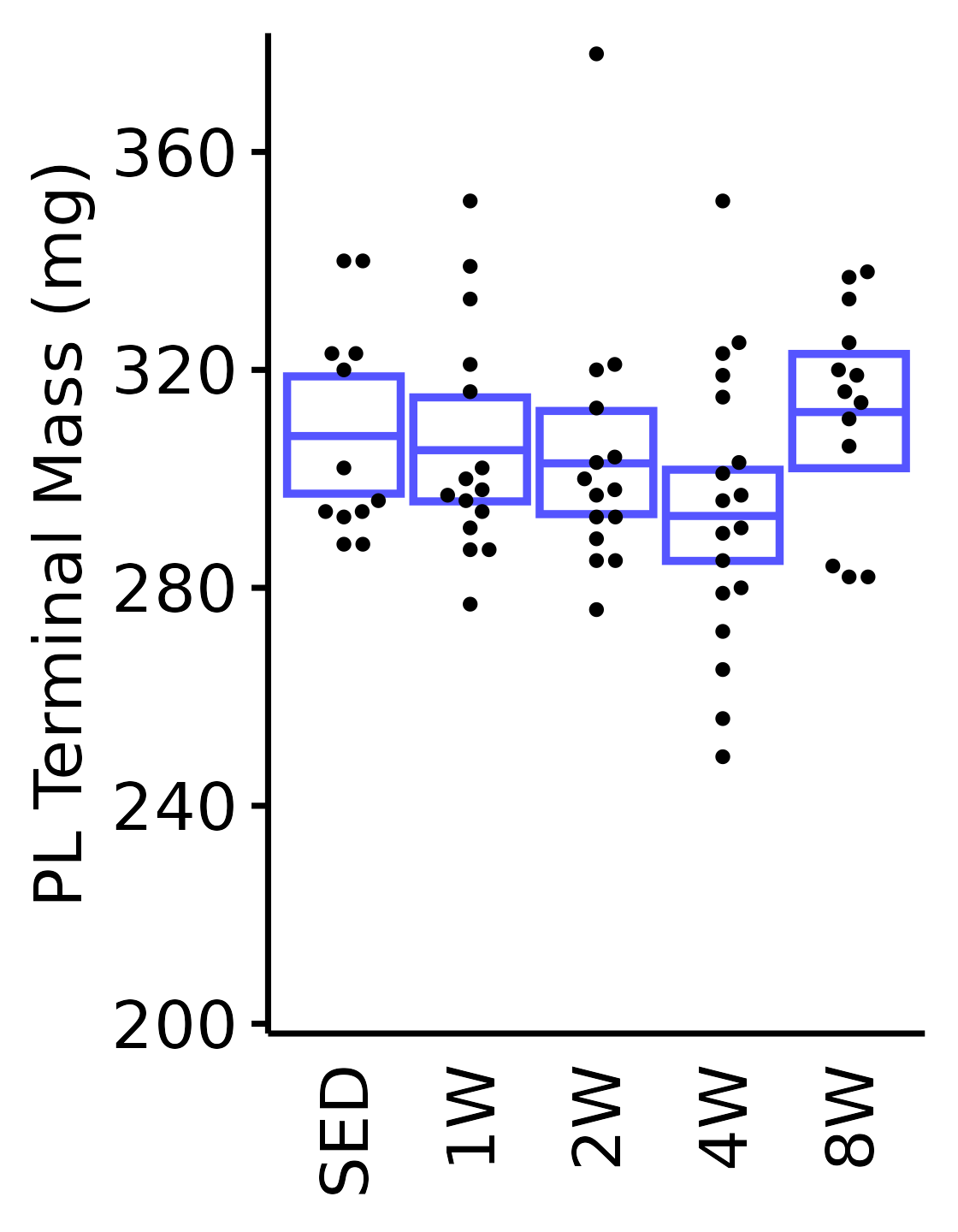
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "PL"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "PL Terminal Mass (mg)",
breaks = seq(160, 220, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(160, 225), clip = "off") +
theme(plot.margin = unit(c(12, 5, 5, 5), units = "pt"))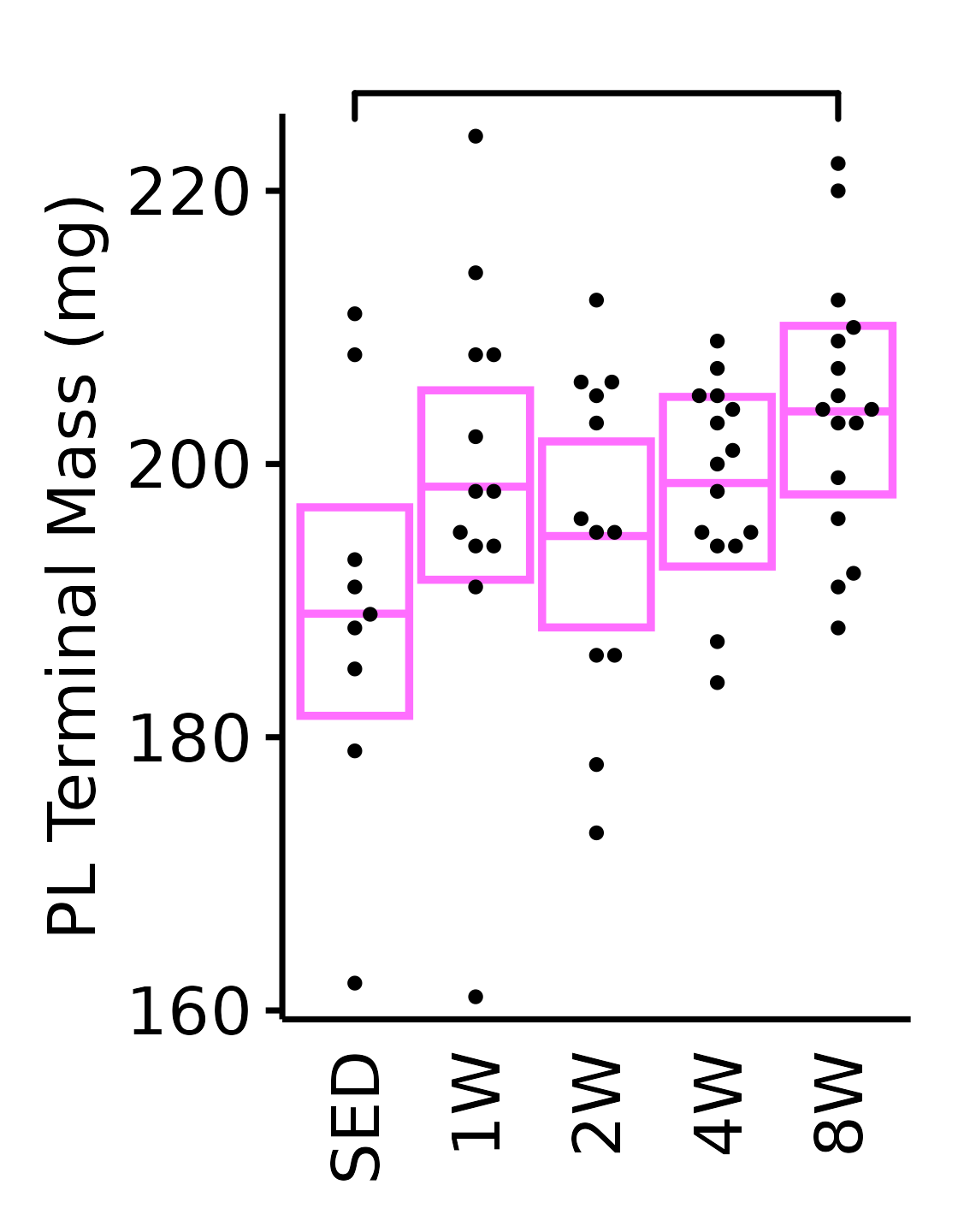
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "PL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "PL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "PL"),
sex = "Male", age = "18M") +
scale_y_continuous(name = "PL Terminal Mass (mg)",
breaks = seq(200, 380, 40),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(200, 380), clip = "off")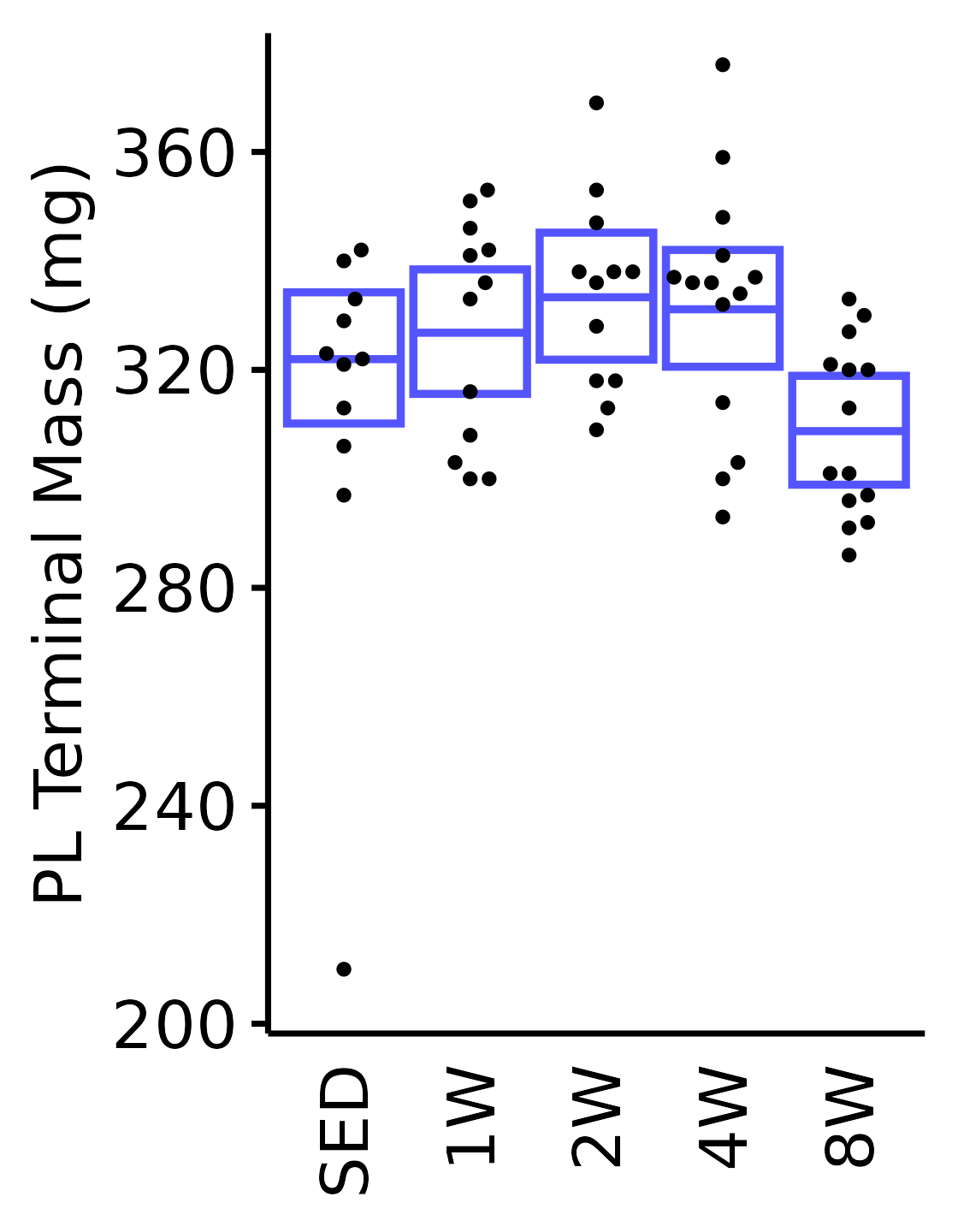
SOL
6M Female
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "SOL"),
sex = "Female", age = "6M", y_position = 101) +
scale_y_continuous(name = "SOL Terminal Mass (mg)",
breaks = seq(60, 110, 10),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(60, 110), clip = "off") +
theme(plot.margin = unit(c(8, 5, 5, 5), units = "pt"))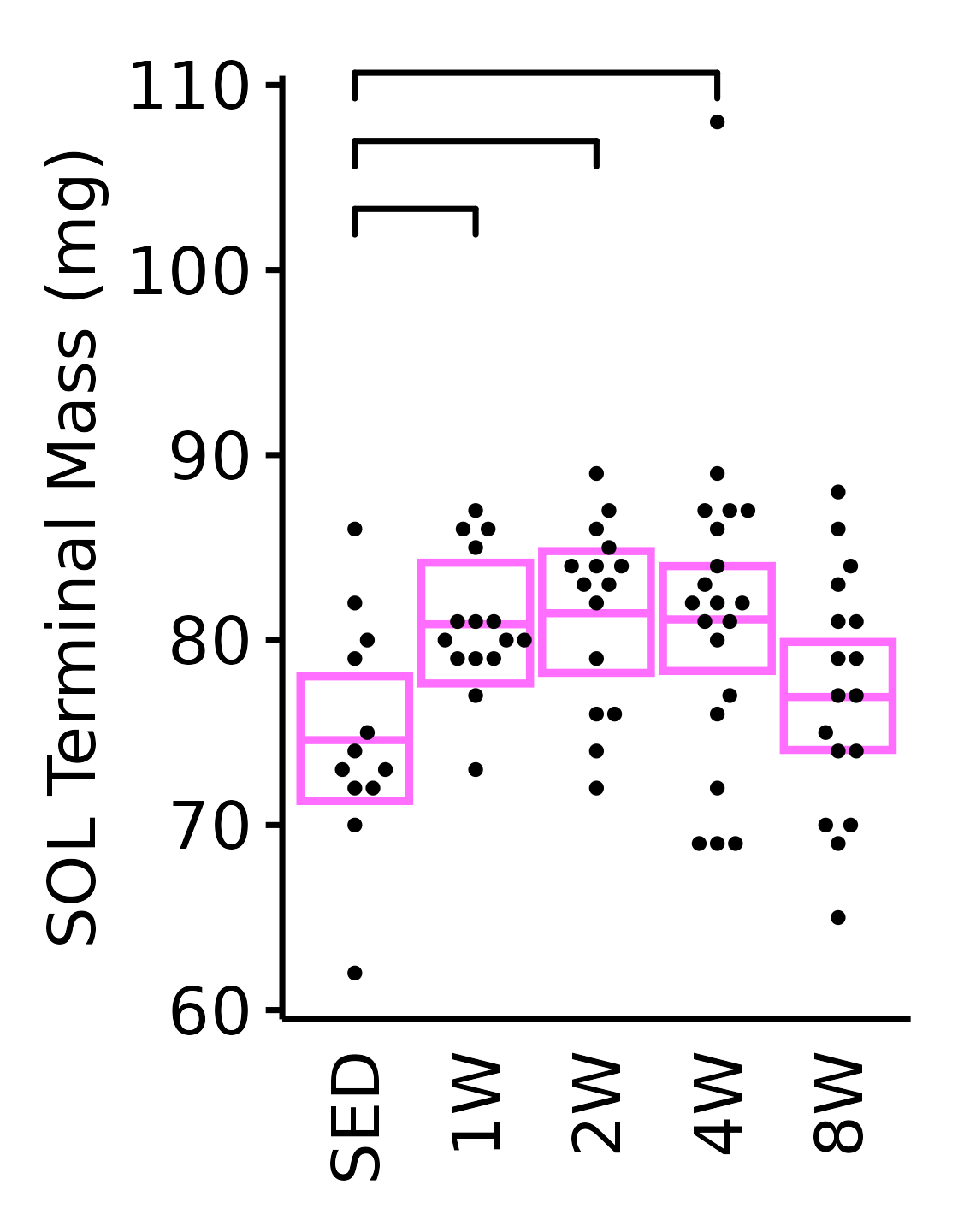
6M Male
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "SOL"),
sex = "Male", age = "6M") +
scale_y_continuous(name = "SOL Terminal Mass (mg)",
breaks = seq(80, 180, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(70, 180), clip = "off")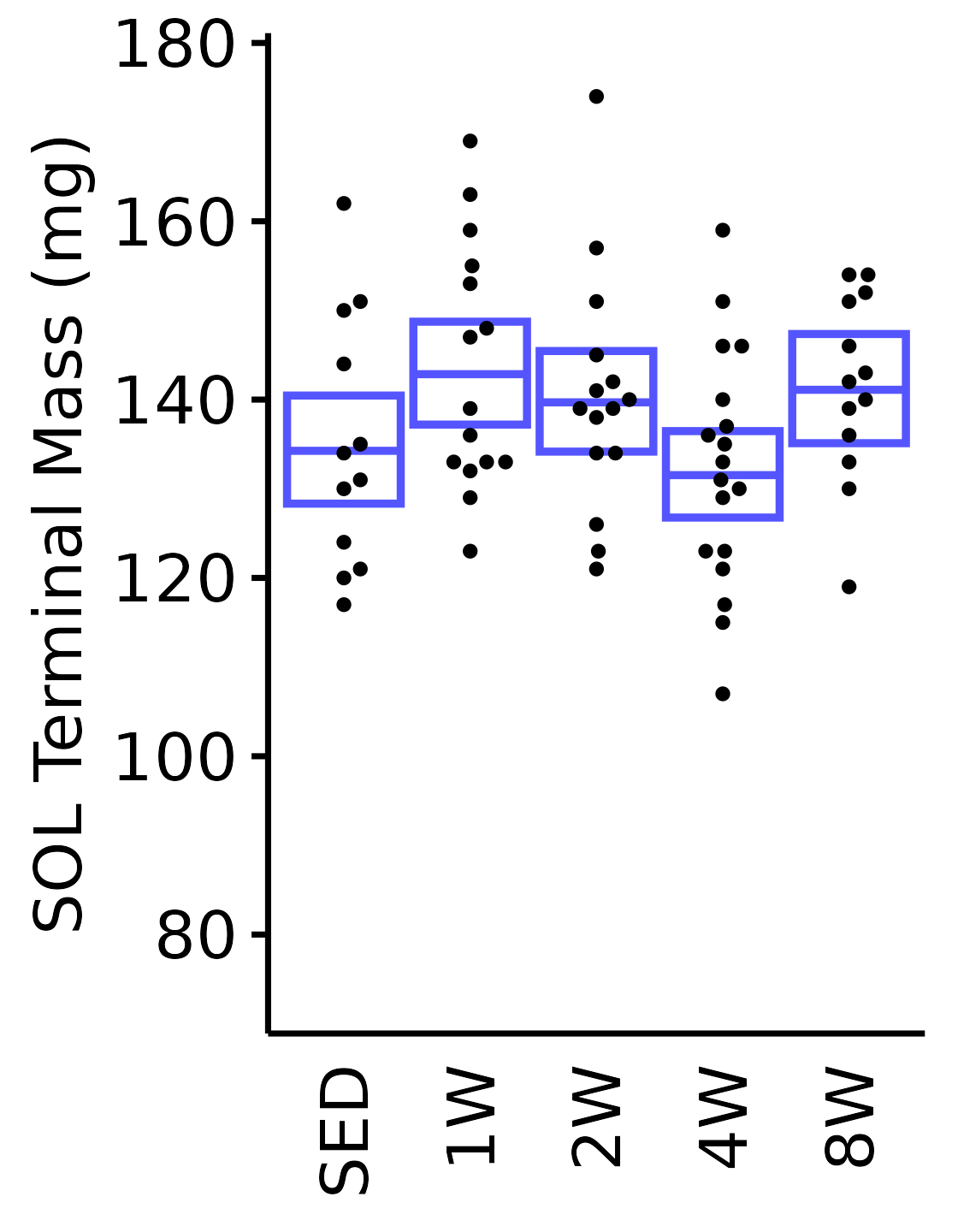
18M Female
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "SOL"),
sex = "Female", age = "18M") +
scale_y_continuous(name = "SOL Terminal Mass (mg)",
breaks = seq(60, 110, 10),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(60, 110), clip = "off") +
theme(plot.margin = unit(c(8, 5, 5, 5), units = "pt"))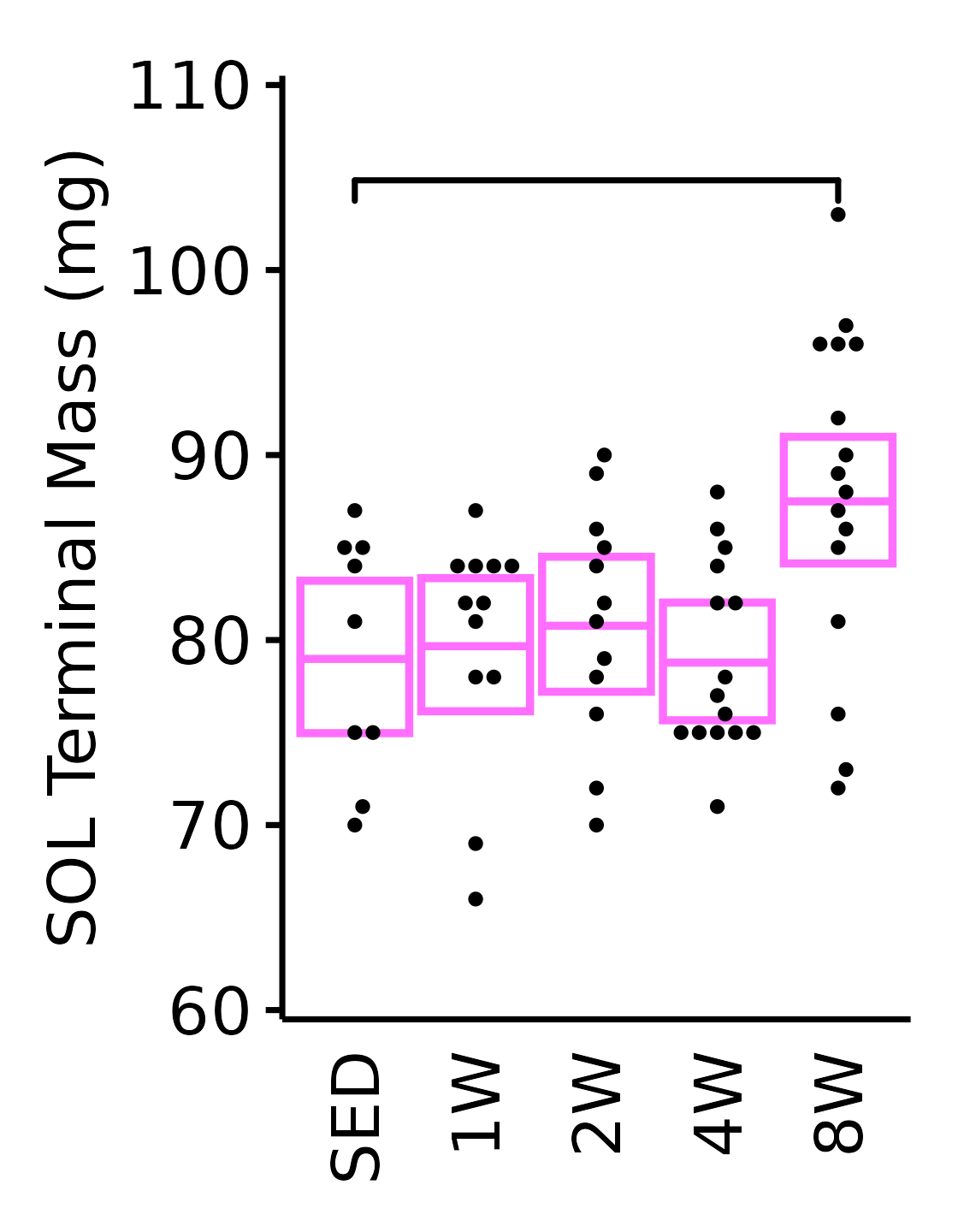
18M Male
plot_baseline(x = filter(MUSCLES, muscle == "SOL"),
response = "term_muscle_mass",
conf = filter(conf_df, response == "Terminal Muscle Mass",
muscle == "SOL"),
stats = filter(MUSCLES_STATS, response == "Terminal Muscle Mass",
muscle == "SOL"),
sex = "Male", age = "18M") +
scale_y_continuous(name = "SOL Terminal Mass (mg)",
breaks = seq(80, 180, 20),
expand = expansion(mult = 1e-2)) +
coord_cartesian(ylim = c(70, 180), clip = "off")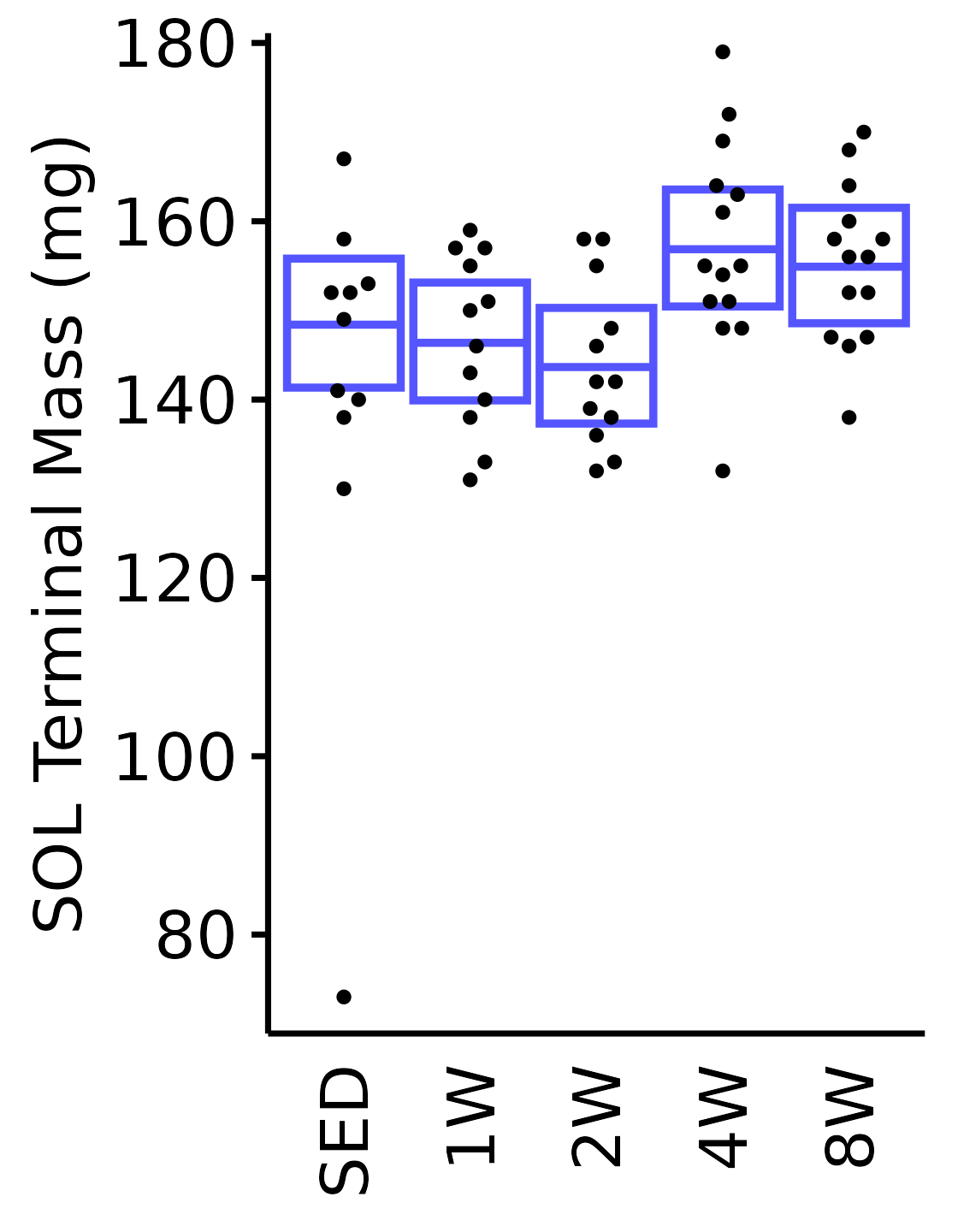
Mean Cross-Sectional Area
Mean muscle cross-sectional area (\(\mu m^2\)).
# Function to plot mean CSA and capillary contacts
plot_function <- function(response,
sex,
age,
conf_df,
stats_df,
ymin,
ymax) {
x <- MUSCLES %>%
dplyr::rename(response = !!sym(response)) %>%
filter(!is.na(response)) %>%
droplevels.data.frame() %>%
filter(age == !!age, sex == !!sex)
conf_df <- filter(conf_df, age == !!age, sex == !!sex)
p <- ggplot(x, aes(x = group, y = response)) +
geom_crossbar(data = conf_df,
aes(x = group, y = response_mean,
ymin = lower.CL, ymax = upper.CL,
color = sex),
fatten = 1, linewidth = 0.5, width = 0.7) +
geom_point(shape = 16, size = 0.5,
position = ggbeeswarm::position_beeswarm(cex = 6,
dodge.width = 0.7))
# Y-axis limits
ylims <- layer_scales(p)$y$range$range
if (missing(ymin))
ymin <- ylims[1]
if (missing(ymax))
ymax <- ylims[2]
# y_pos <- x %>%
# group_by(muscle) %>%
# summarise(y.position = max(response))
y_position <- x %>%
left_join(select(conf_df, muscle, group, upper.CL),
by = c("group", "muscle")) %>%
group_by(muscle) %>%
summarise(y.position = max(c(response, upper.CL), na.rm = TRUE) +
0.1 * (ymax - ymin))
stats_df <- stats_df %>%
filter(signif) %>%
filter(age == !!age, sex == !!sex) %>%
mutate(contrast = as.character(contrast),
group1 = sub(" .*", "", contrast),
group2 = sub(".* ", "", contrast)) %>%
left_join(y_position, by = "muscle")
p <- p +
ggpubr::stat_pvalue_manual(data = stats_df,
label = NULL,
size = 0, # hide asterisks
bracket.nudge.y = 0,
bracket.size = 0.6) +
annotation_custom(grob = linesGrob(x = c(0, 1),
y = unit(-20, "pt"),
gp = gpar(lwd = 0.9)),
xmin = -Inf, xmax = Inf,
ymin = -Inf, ymax = Inf)
## Modify appearance
p <- p +
facet_grid(~ muscle, switch = "x") +
scale_color_manual(values = c("#ff6eff", "#5555ff"),
breaks = c("Female", "Male")) +
labs(x = NULL) +
coord_cartesian(ylim = c(ymin, ymax), clip = "off") +
theme_bw(base_size = 7) +
theme(text = element_text(size = 7, color = "black"),
line = element_line(linewidth = 0.3, color = "black"),
axis.ticks = element_line(linewidth = 0.3, color = "black"),
panel.grid = element_blank(),
panel.border = element_blank(),
axis.ticks.x = element_blank(),
axis.text = element_text(size = 7,
color = "black"),
axis.text.x = element_text(size = 7, angle = 90, hjust = 1,
vjust = 0.5),
axis.title = element_text(size = 7, margin = margin(),
color = "black"),
axis.line = element_line(color = "black", linewidth = 0.3),
strip.background = element_blank(),
strip.text = element_text(size = 7, color = "black",
margin = margin(b = 0, t = 2)),
panel.spacing = unit(2, "pt"),
plot.title = element_text(size = 8, color = "black"),
plot.subtitle = element_text(size = 7, color = "black"),
legend.position = "none",
strip.placement = "outside"
)
return(p)
}6M Female
plot_function(
response = "mean_CSA", sex = "Female", age = "6M",
conf_df = filter(conf_df, response == "Mean CSA"),
stats_df = filter(MUSCLES_STATS, response == "Mean CSA"),
ymin = 1000, ymax = 2500) +
scale_y_continuous(name = TeX("Mean CSA ($\\mu m^2$)"),
breaks = seq(1000, 2500, 500),
expand = expansion(mult = 5e-3))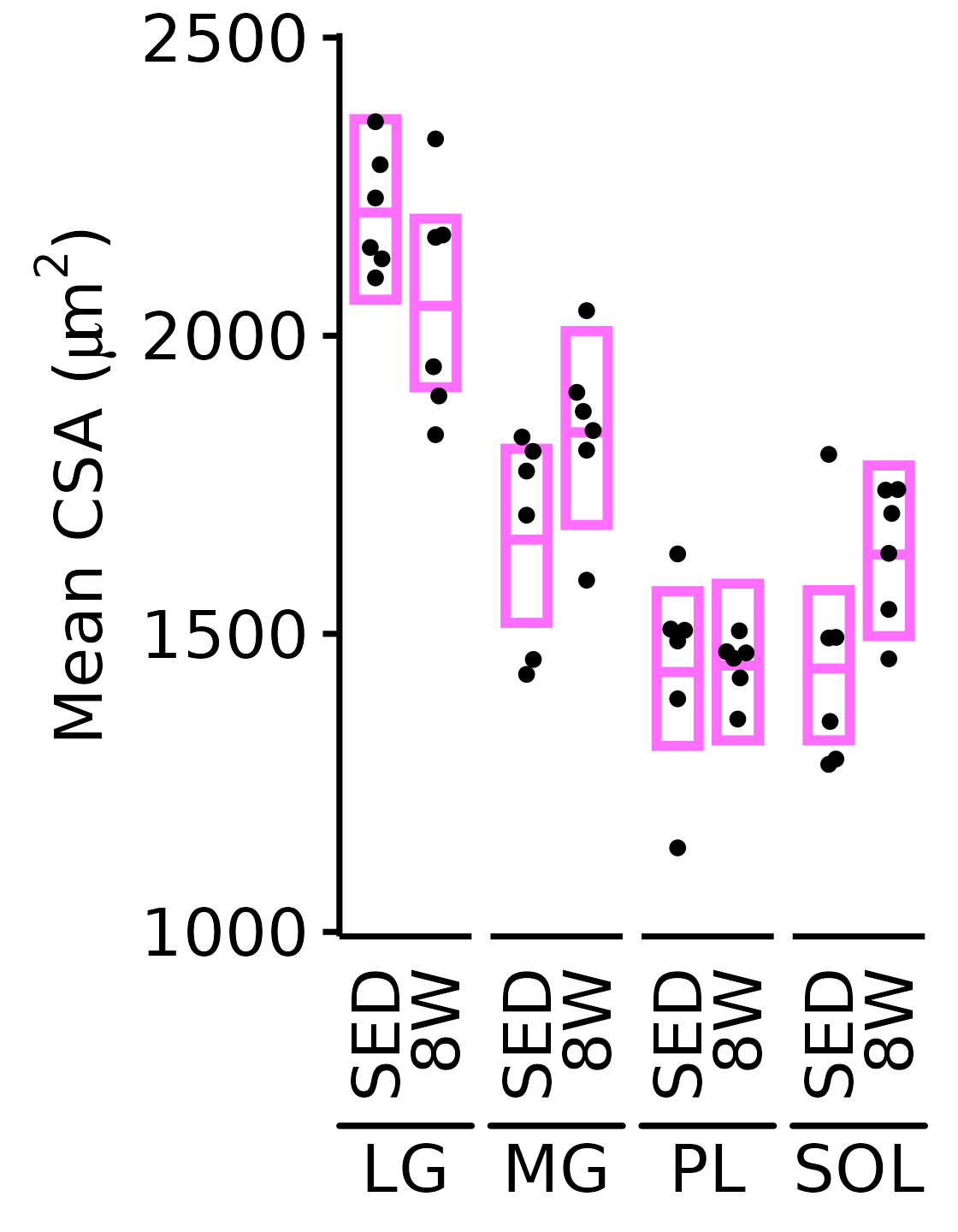
6M Male
plot_function(
response = "mean_CSA", sex = "Male", age = "6M",
conf_df = filter(conf_df, response == "Mean CSA"),
stats_df = filter(MUSCLES_STATS, response == "Mean CSA"),
ymin = 1500, ymax = 3500) +
scale_y_continuous(name = TeX("Mean CSA ($\\mu m^2$)"),
breaks = seq(1500, 3500, 500),
expand = expansion(mult = 5e-3))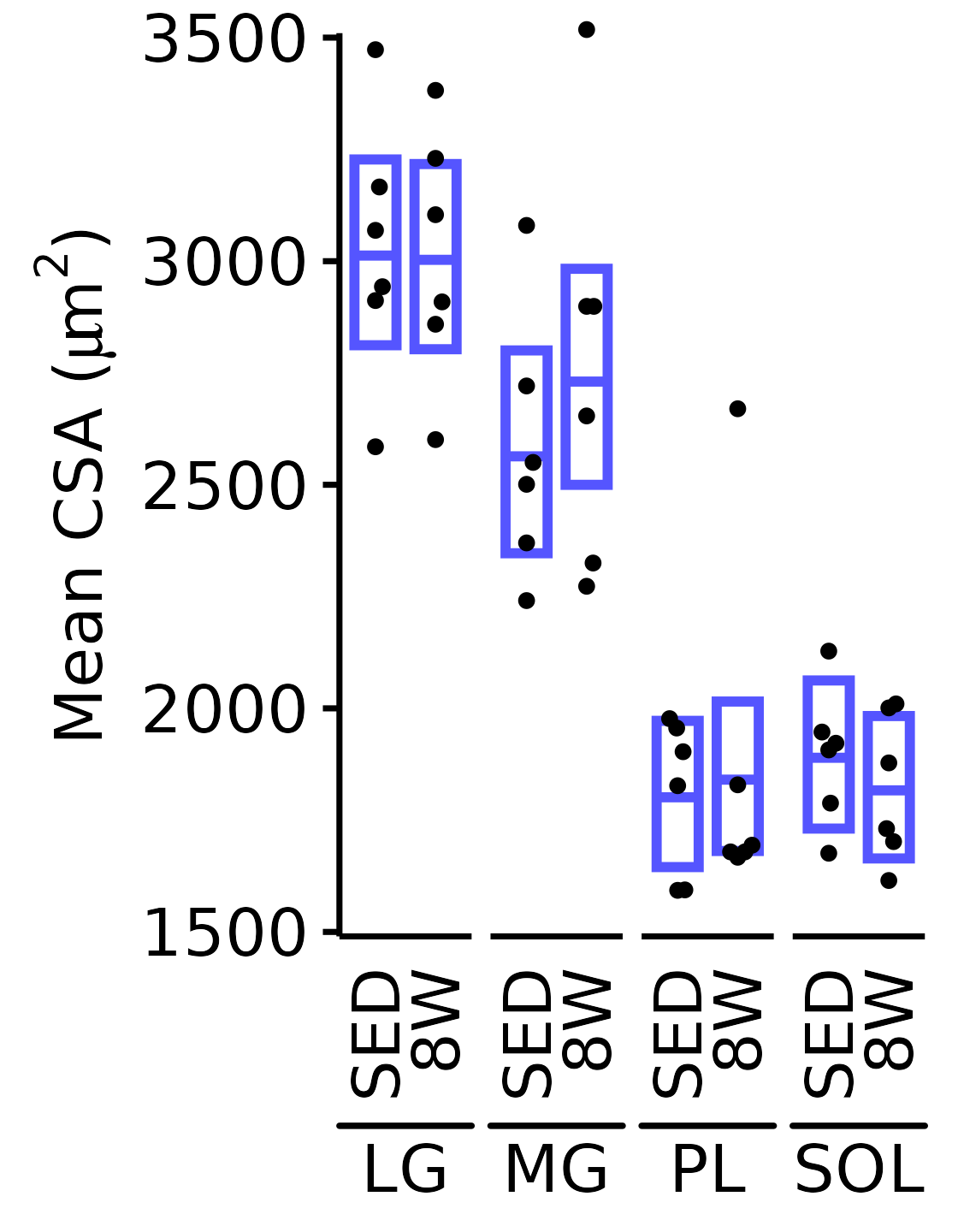
18M Female
plot_function(
response = "mean_CSA", sex = "Female", age = "18M",
conf_df = filter(conf_df, response == "Mean CSA"),
stats_df = filter(MUSCLES_STATS, response == "Mean CSA"),
ymin = 1000, ymax = 2500) +
scale_y_continuous(name = TeX("Mean CSA ($\\mu m^2$)"),
breaks = seq(1000, 2500, 500),
expand = expansion(mult = 5e-3))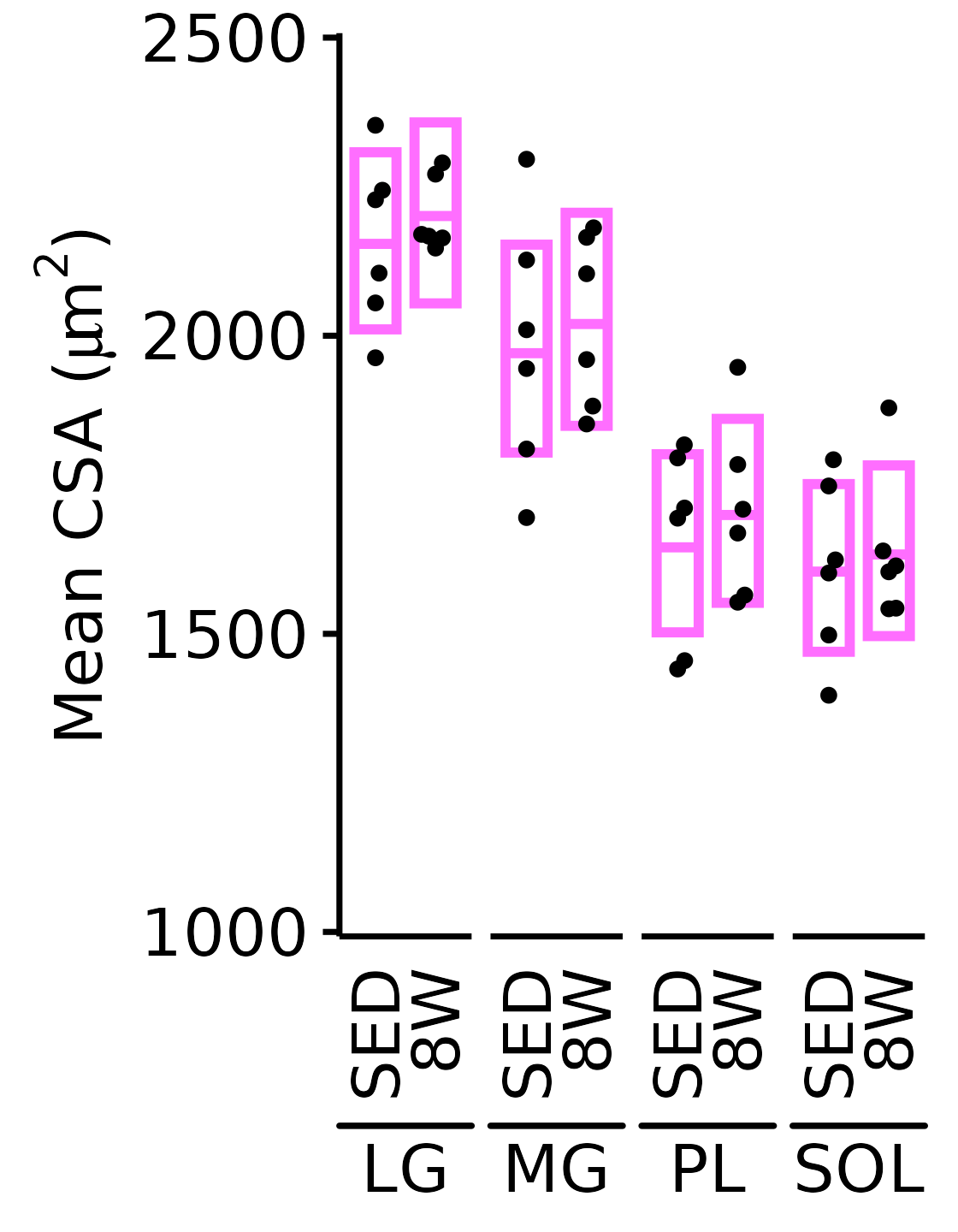
18M Male
plot_function(
response = "mean_CSA", sex = "Male", age = "18M",
conf_df = filter(conf_df, response == "Mean CSA"),
stats_df = filter(MUSCLES_STATS, response == "Mean CSA"),
ymin = 1500, ymax = 3500) +
scale_y_continuous(name = TeX("Mean CSA ($\\mu m^2$)"),
breaks = seq(1500, 3500, 500),
expand = expansion(mult = 5e-3))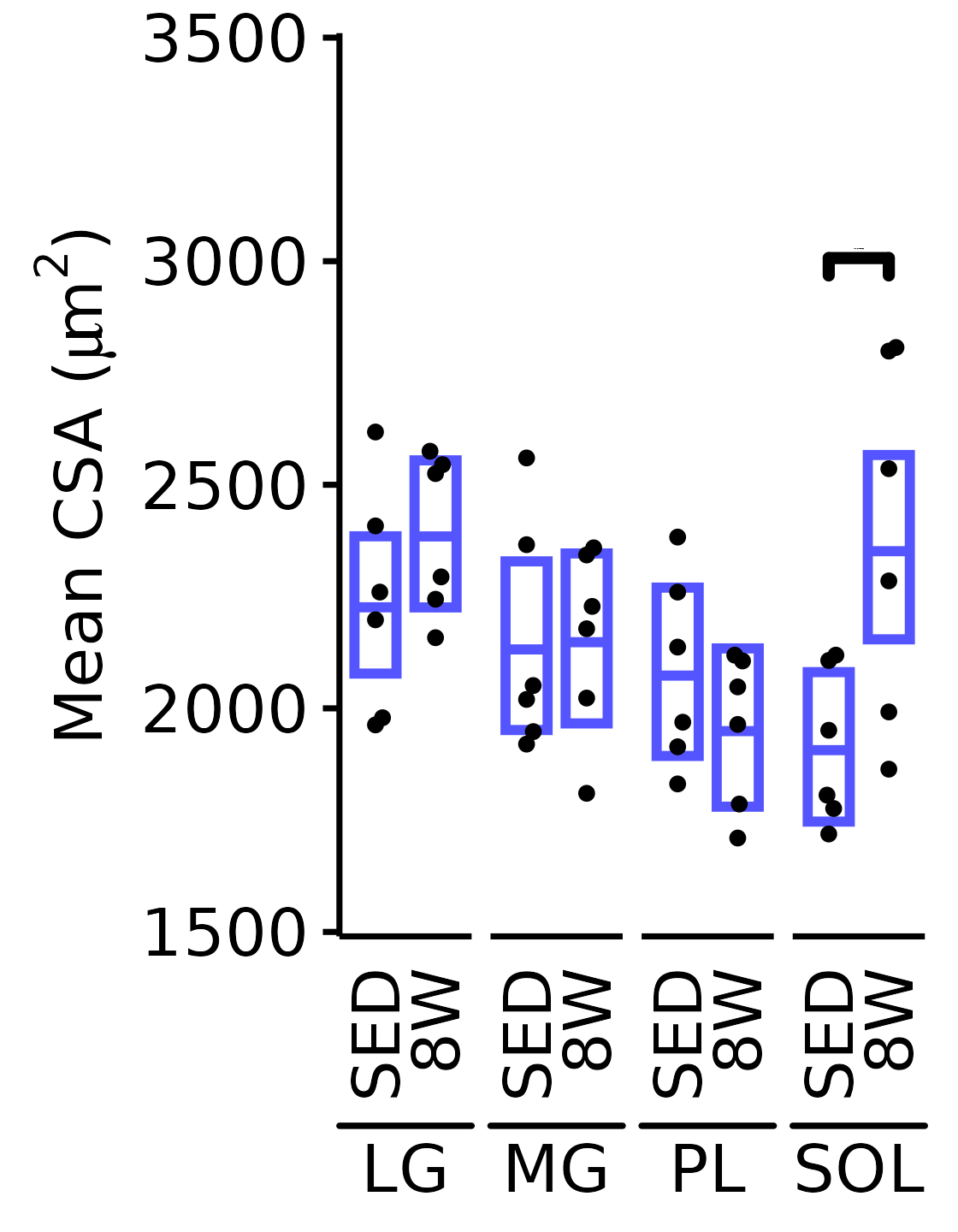
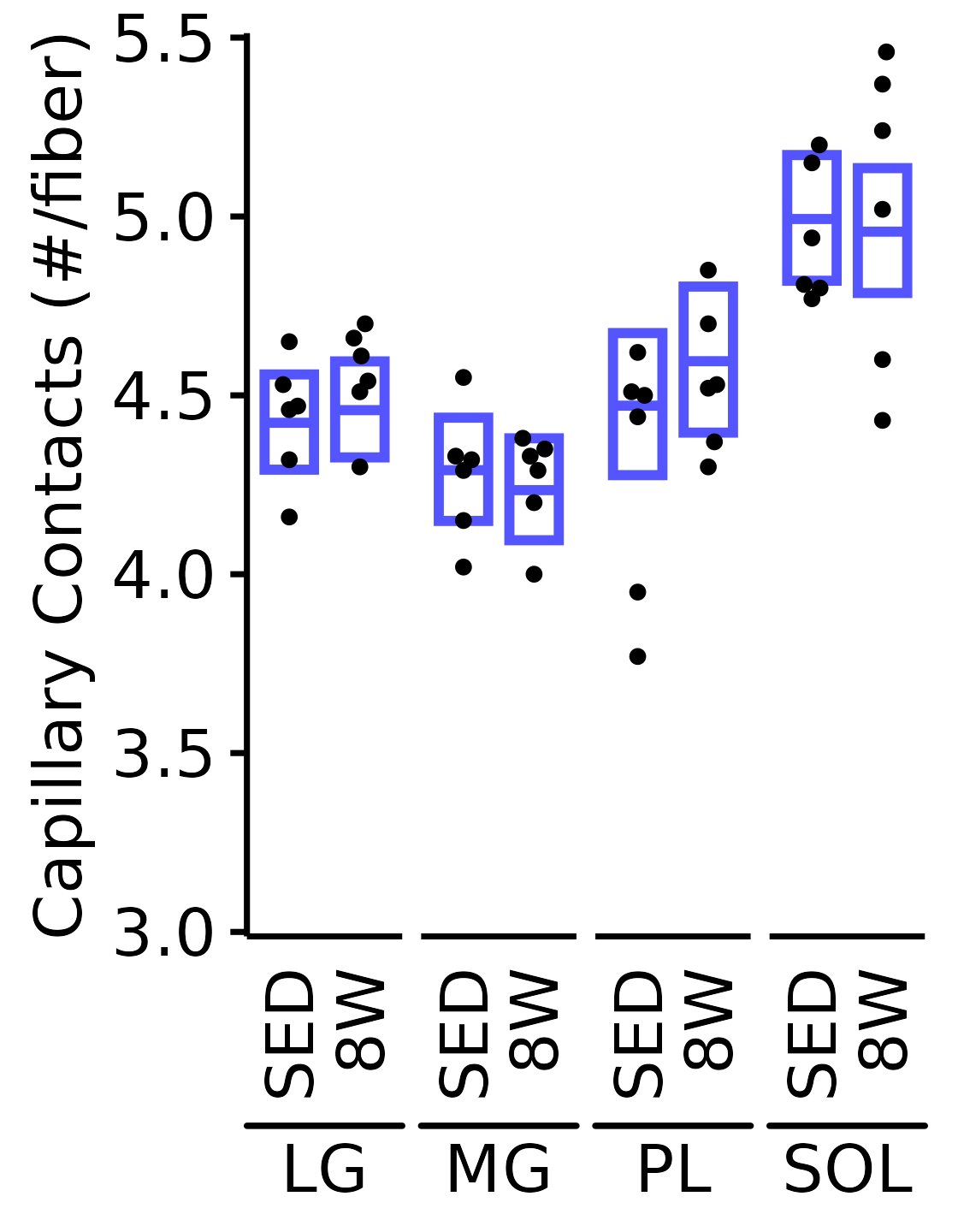
Capillary Contacts
Mean number of muscle capillary contacts (capillaries/fiber).
6M Female
plot_function(
response = "capillary_contacts", sex = "Female", age = "6M",
conf_df = filter(conf_df, response == "Capillary Contacts"),
stats_df = filter(MUSCLES_STATS, response == "Capillary Contacts"),
ymin = 3, ymax = 5.5) +
scale_y_continuous(name = "Capillary Contacts (#/fiber)",
breaks = seq(3, 5.5, 0.5),
expand = expansion(mult = 5e-3))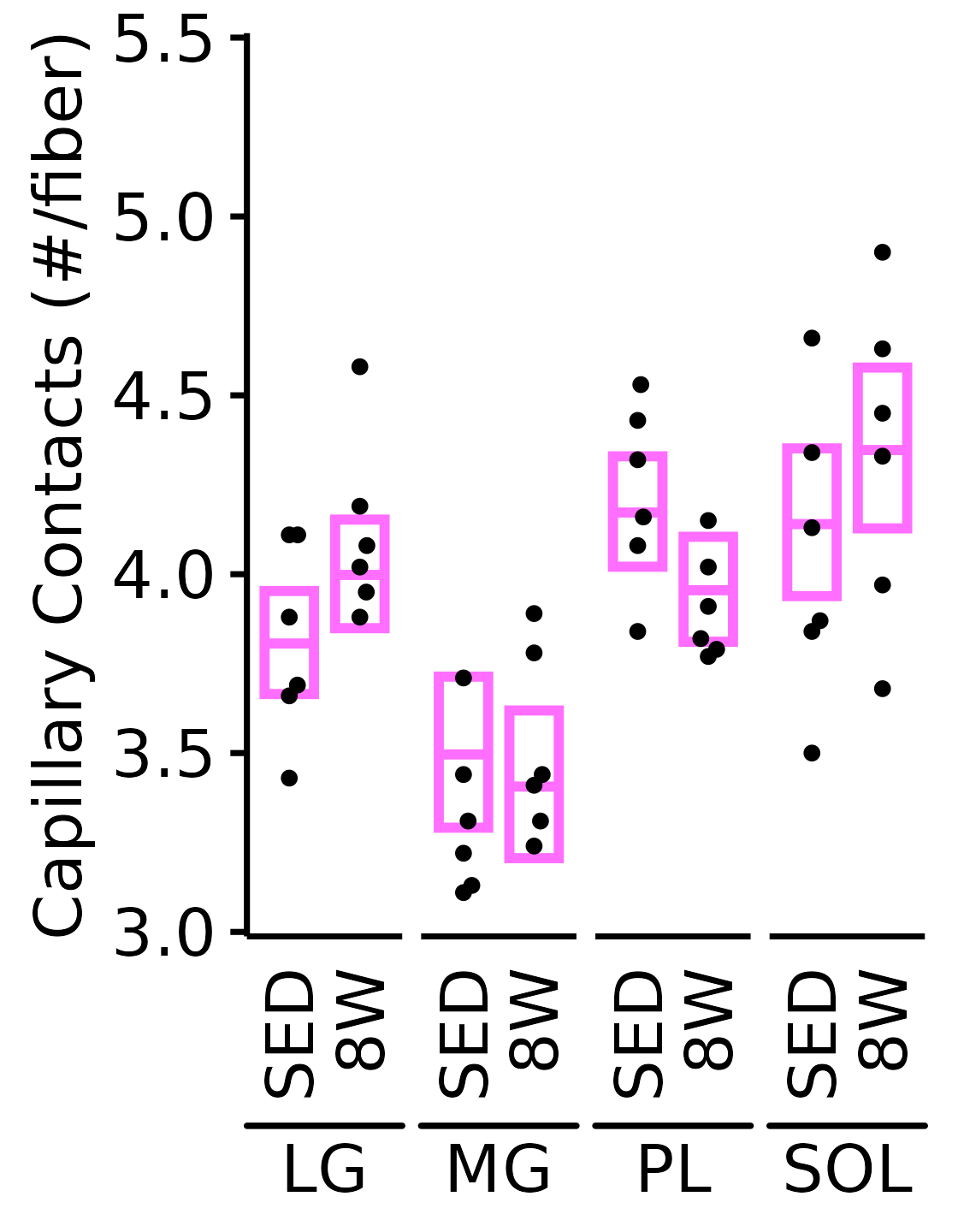
6M Male
plot_function(
response = "capillary_contacts", sex = "Male", age = "6M",
conf_df = filter(conf_df, response == "Capillary Contacts"),
stats_df = filter(MUSCLES_STATS, response == "Capillary Contacts"),
ymin = 3, ymax = 5.5) +
scale_y_continuous(name = "Capillary Contacts (#/fiber)",
breaks = seq(3, 5.5, 0.5),
expand = expansion(mult = 5e-3))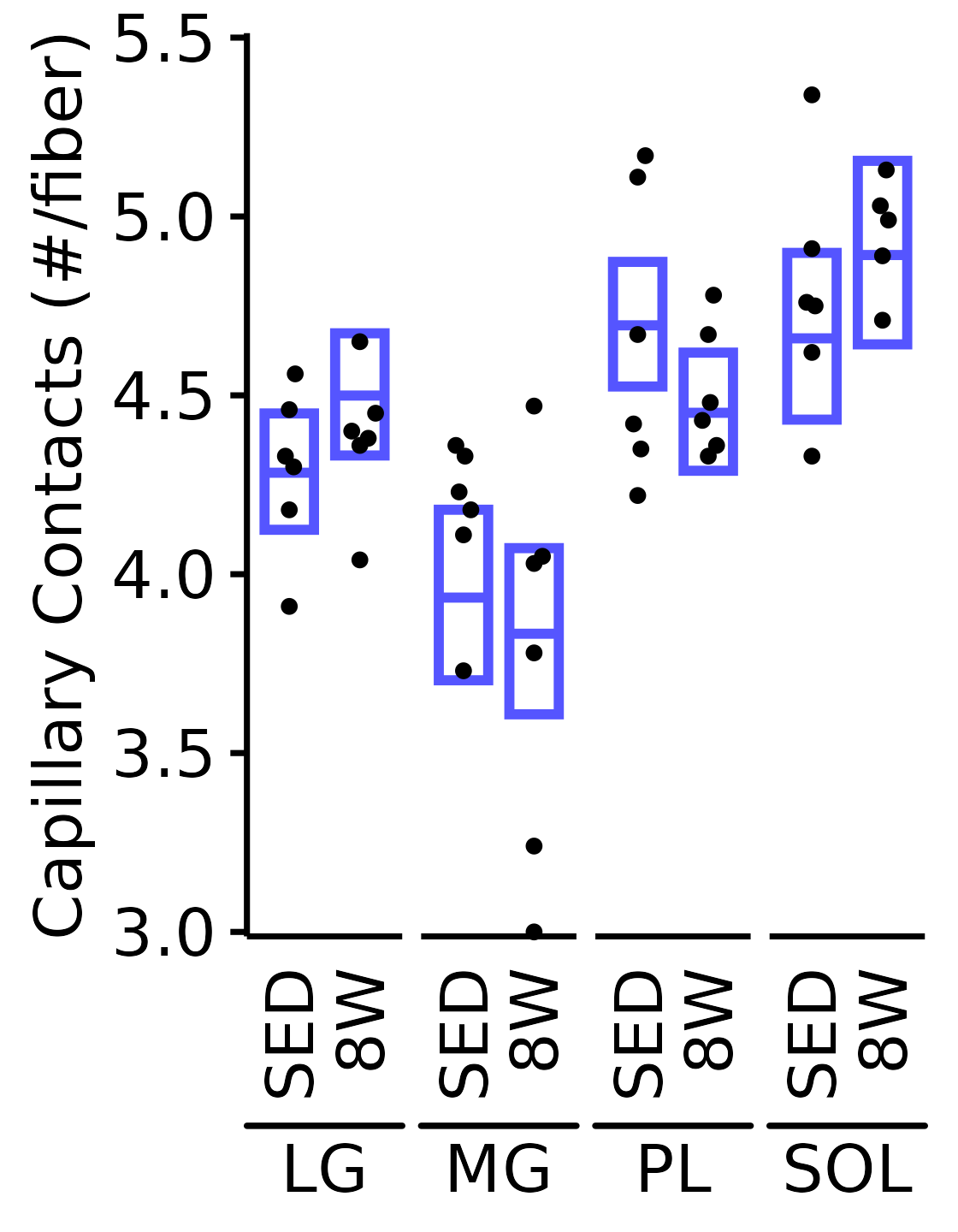
18M Female
plot_function(
response = "capillary_contacts", sex = "Female", age = "18M",
conf_df = filter(conf_df, response == "Capillary Contacts"),
stats_df = filter(MUSCLES_STATS, response == "Capillary Contacts"),
ymin = 3, ymax = 5.5) +
scale_y_continuous(name = "Capillary Contacts (#/fiber)",
breaks = seq(3, 5.5, 0.5),
expand = expansion(mult = 5e-3))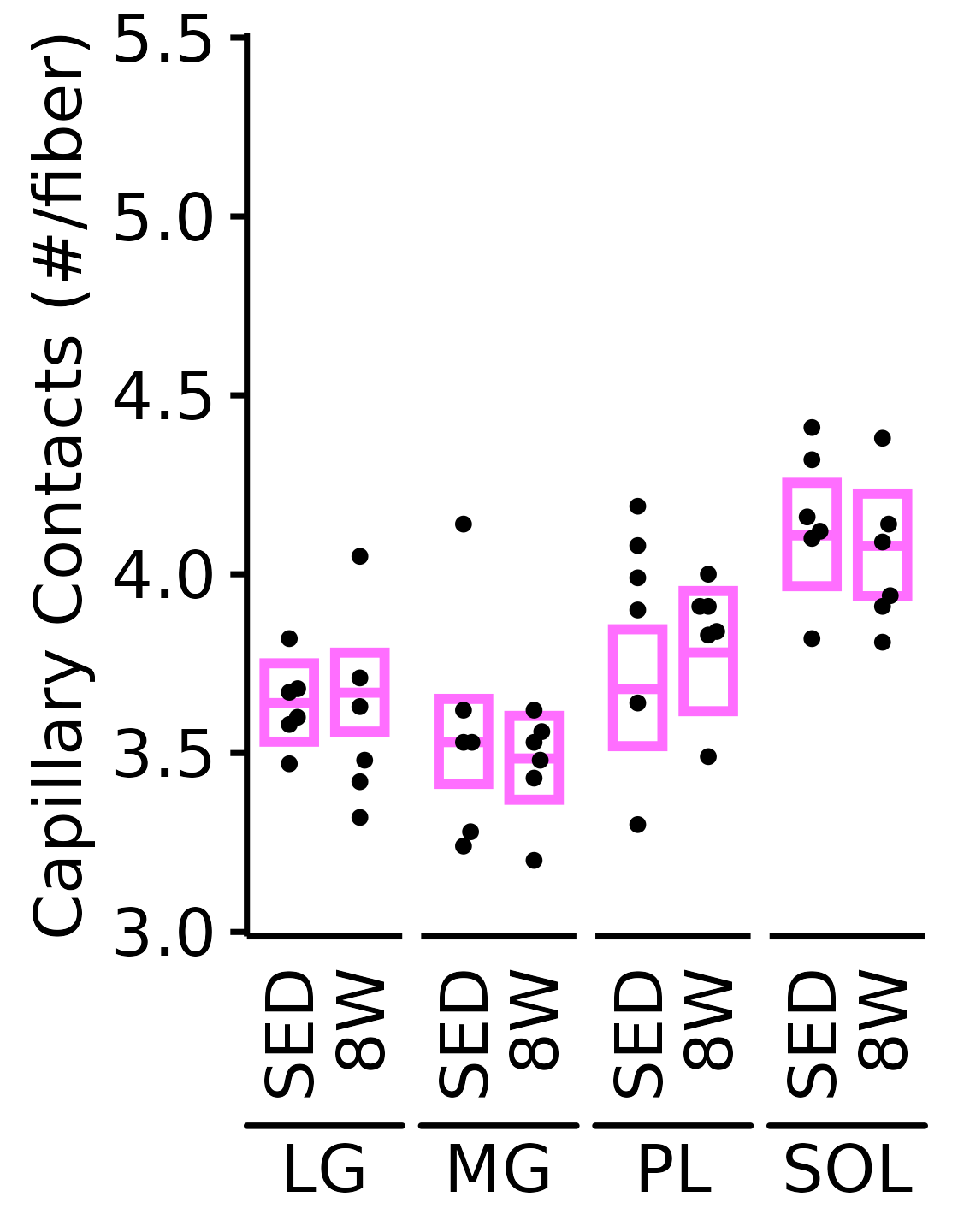
18M Male
plot_function(
response = "capillary_contacts", sex = "Male", age = "18M",
conf_df = filter(conf_df, response == "Capillary Contacts"),
stats_df = filter(MUSCLES_STATS, response == "Capillary Contacts"),
ymin = 3, ymax = 5.5) +
scale_y_continuous(name = "Capillary Contacts (#/fiber)",
breaks = seq(3, 5.5, 0.5),
expand = expansion(mult = 5e-3))